Figure 5.
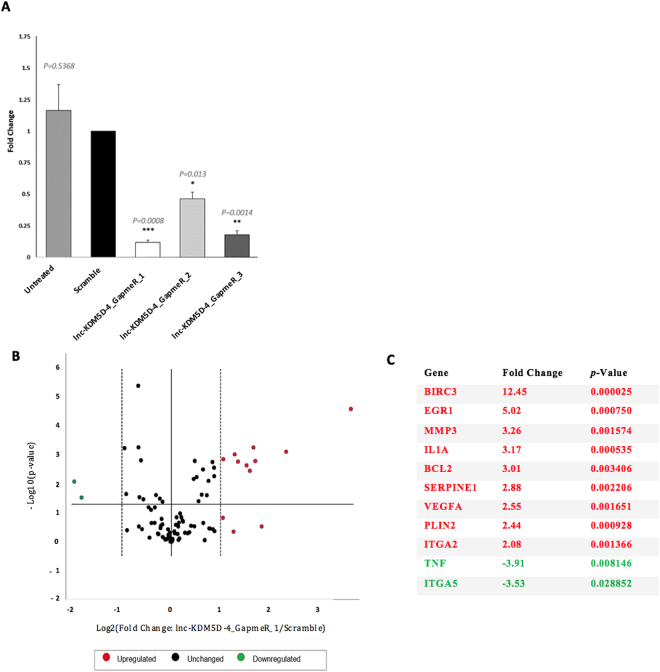
Lnc-KDM5D-4 knockdown and associated atherosclerosis-relevant genes. Cells were transfected with Locked Nucleic Acid (LNA) GapmeRs targeting lnc-KDM5D-4:1 RNA or random scrambled sequence. (A) Cells were harvested 48 h after transfection. AO-mediated silencing of lnc-KDM5D-4 was assessed in real-time PCR (normalised to ACTB as housekeeping control gene). The cells demonstrate antisense oligonucleotide knockdown of nuclear-located lnc-KDM5D-4 with the 3-different tested GapmeRs; a highest knockdown efficiency was achieved with the lnc-KDM5D-4_GapmeR_1 [transcript reduction by 88% (Fold change = 0.12; p-value = 0.0008)]. Comparison of transfected cells with GapmeRs, and Scramble to Untreated samples (negative control) showed that there was no significant effect of transfection reagent or transfection reagent plus GapmeR respectively on the cell. (B,C) A Human Atherosclerosis RT2 Profiler™ PCR Array was utilized to profile the expression of 84 genes involved in atherosclerosis between HepG2 cells transfected with GapmeR targeting lnc-KDM5D-4 (lnc-KDM5D-4_GapmeR_1) and scrambled cells. Relative expression levels of these genes were normalised to a set of housekeeping genes (Supplementary Table S6). Three independent biological samples were used (n = 3). The volcano plot combines a p-value statistical test with the fold change enabling identification of genes with both large and small expression changes that are statistically significant are represented; upregulated genes (red), downregulated genes (green), and unchanged genes (black) (B). In transfected cells compared to untransfected cells, results determine that 9 genes were significantly over-expressed ((p-value < 0.05; Fold Change >2): BIRC3 (baculoviral IAP repeat containing 3), EGR1 (early growth response 1), MMP3 (matrix metallopeptidase 3), IL1A (interleukin 1 alpha), BCL2 (BCL2, apoptosis regulator), SERPINE1 (serpin family E member 1), VEGFA (vascular endothelial growth factor A), PLIN2 (perilipin 2), and ITGA2 (integrin subunit alpha 2); and 2 genes were significantly under-expressed ((p-value < 0.05; Fold Change <−2.00): ITGA5 (integrin subunit alpha 5), and TNF (tumor necrosis factor) (C). The significant dysregulated genes are summarized in Supplementary Figure S2 with their pathways associated with the most represented ‘anti-apoptosis pathway’. Statistical significance: *p-value < 0.05; **p-value < 0.01; ***p-value < 0.001.