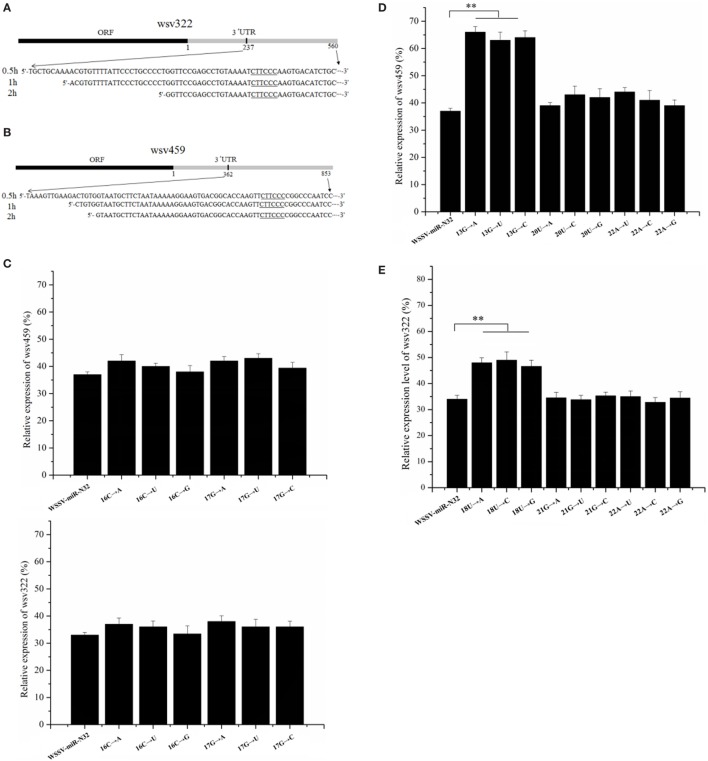
Figure 5.
Sites of a microRNA (miRNA) required for miRNA targeting. (A,B) The miRNA-guided cleavage of target mRNA. White spot syndrome virus (WSSV)-miR-N32 was incubated with the Ago1 complex and 3′ untranslated region (3′UTR) of wsv322 mRNA (A) or 3′UTR of wsv459 mRNA (B) for different time as indicated on the left. Then, the cleaved fragment was sequenced. The sequence complementary to the seed sequence of WSSV-miR-N32 was underlined. (C) The influence of uncomplementary bases of WSSV-miR-N32 non-seed sequence on the expression of its target genes. The mutated or wild-type WSSV-miR-N32 was co-injected with WSSV into shrimp. The shrimp were cultured for 36 h. Then, the shrimp hemocytes were subjected to quantitative real-time polymerase chain reaction (PCR) to detect wsv322 mRNA or wsv459 mRNA. The arrows represented the mutated sites and bases of WSSV-miR-N32. (D,E) The effects of base mutation of WSSV-miR-N32 non-seed sequence on the expression of its target genes. WSSV and wild-type or mutated WSSV-miR-N32 were injected into shrimp. Thirty-six hours later, the wsv322 mRNA level (D) or wsv459 mRNA level (E) in shrimp hemocytes was examined with quantitative real-time PCR. The arrows indicated the mutated sites and bases of WSSV-miR-N32. In all panels, the statistically significant differences between treatments were shown with asterisks (**p < 0.01).