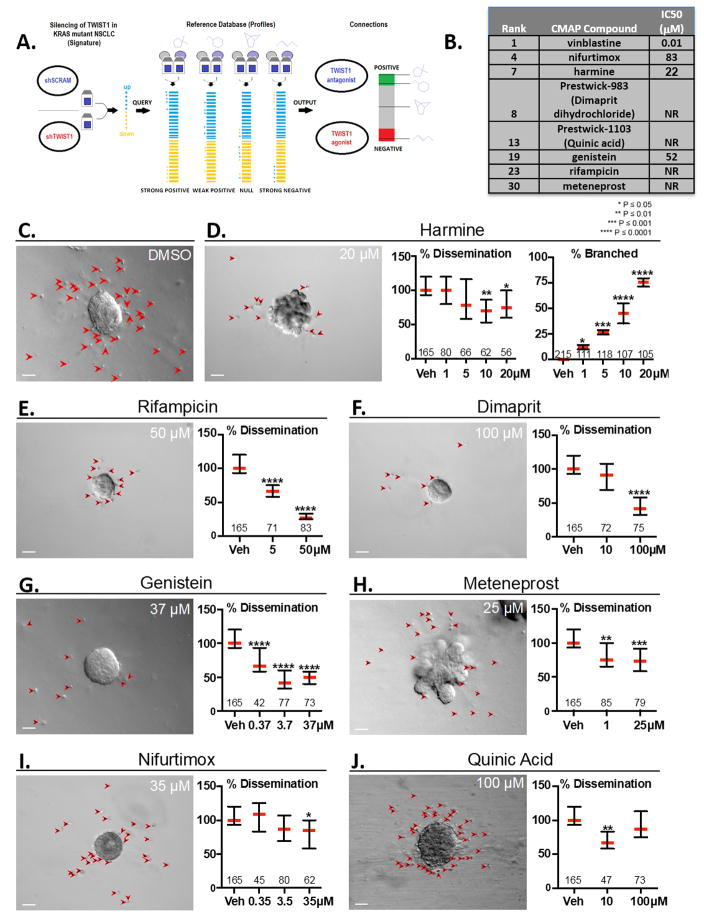
Figure 1. Connectivity MAP (CMAP) analysis identifies compounds which inhibit NSCLC proliferation, TWIST1-dependent dissemination and TWIST1-suppression of organoid branching.
(A) CMAP Analysis Schematic. A gene signature for TWIST1 knockdown was utilized to query the Connectivity Map (Broad Institute). (B) Chart depicting eight of the top 30 ranked compounds which were selected for further analysis. IC50 was determined in two KRAS mutant NSCLC cell lines (A549 and H460) at 72 hours using MTS assays. (C) TWIST1 induction with doxycycline results in profound dissemination of cells and prevents branching (DMSO, upper left panel). Primary Twist1 inducible breast epithelial cells are implanted as organoids in 3D culture. Red arrowheads indicate disseminated cells. (D–J) Candidate compounds inhibit Twist1-induced 3D dissemination and/or restore FGF2-induced branching of primary breast epithelial cells in vitro. Values within the graph indicate the number of organoids quantified per treatment condition. Dissemination data is normalized to the median of each experimental replicate. Error bars, 95% Confidence Intervals. Treatment with small molecules listed at the indicated doses induced statistically significant (Kruskal-Wallis test, P<.05) reductions in dissemination. (D) Branching data is presented as mean of each experimental replicate. See Supplementary Fig. 1A–C for representative images of unbranched vs. branched organoids. Error bars, ±SD. Treatment with harmine at the indicated doses induced statistically significant (One-way ANOVA, P<.05) increases in branching. Scale bar, 50μm.