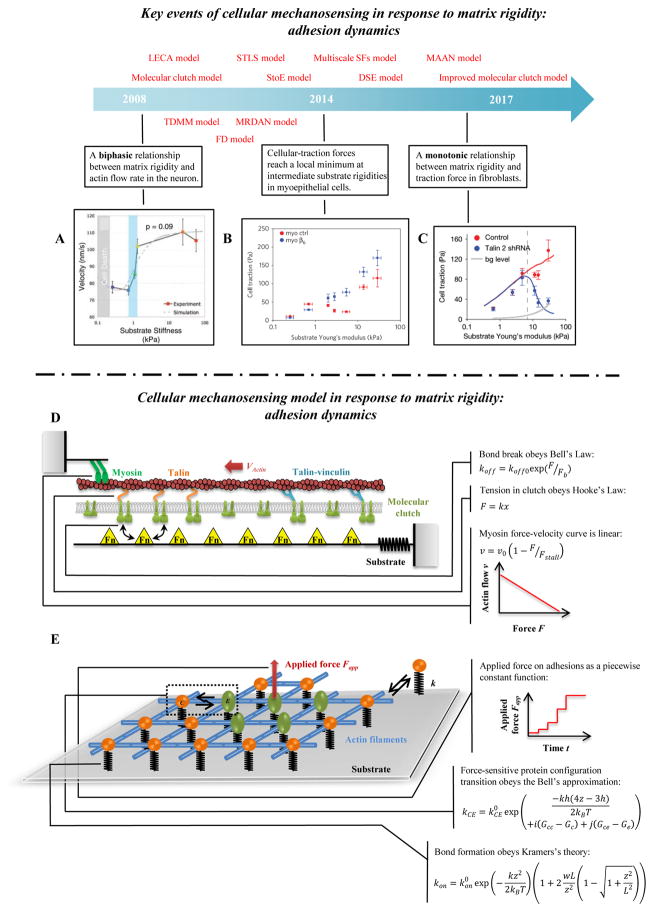
Figure 4. Cellular mechanosensing in response to matrix rigidity: adhesion dynamics.
(A) Neurons have higher actin flow rate on stiffer substrata [147]. (B–C) The distinct actin flow/matrix rigidity relationships in breast myoepithelial cells and fibroblasts [10, 25]. (D) Schematic of the uniaxial molecular clutch model. Actin polymerization and depolymerization at the tips of filopodia are coupled to the substrate through molecular clutches, and these molecular clutches resist the retrograde actin flow driven by myosin motors and membrane fluctuations. With increasing tension, the following transformations of molecular clutches are possible: talin unfolding and refolding, clutch reinforcement by vinculin binding, signal activation from clutch reconfiguration, and weakest-link rupture. (E) Schematic of the 2D molecular-mechanical adhesion model. Each molecule may bind to the substrate through a flexible spring, and may transition from a circular to an elliptical state under mechanical loading.