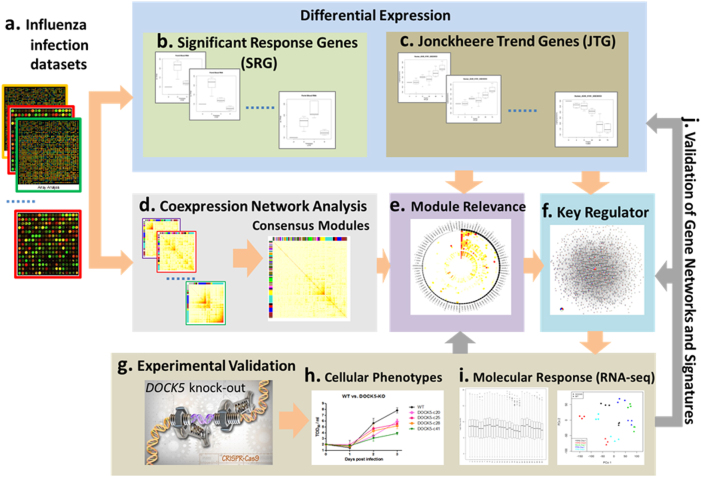
Fig. 1.
Overview of the proposed integrative network-based approach to influenza infection. a Twelve publically available gene expression data sets for studying influenza infection were curated. b ANOVA was used to identify significant response genes (SRG) that were differentially expressed by taking into account the time-series. c Genes showing significant up-regulation or down-regulation along the time-series were identified by the Jonckheere trend analysis and they were called Jonckheere trend genes (JTG). d Weighted co-expression network analysis was applied to each data set to identify modules comprised of highly co-regulated genes. Consensus modules were further determined by the clustering analysis of the similarities between all the modules from the 12 data sets. e The consensus modules were rank-ordered by their enrichment for both SRGs and JTGs. f Key regulators of the top-ranked module were determined by network connectivity. Key driver centered un-weighted co-expression networks were then constructed by correlation analysis of the individual data sets. g Key regulator (DOCK5) predicted by the network analysis was validated through CRISPR-Cas9-based knockout experiments. h The impact of DOCK5 knockout on the replication of the virus in A549 cells was determined by titration of the culture supernatants using a TCID50 assay. i Molecular response to DOCK5 knockout, i.e., DOCK5’s target genes, was determined by RNA sequencing data from the CRISPR-Cas9-based knockout experiments. j The predicted gene networks and differentially expressed gene signatures were validated by their enrichment of DOCK5 target genes identified through the validation experiments