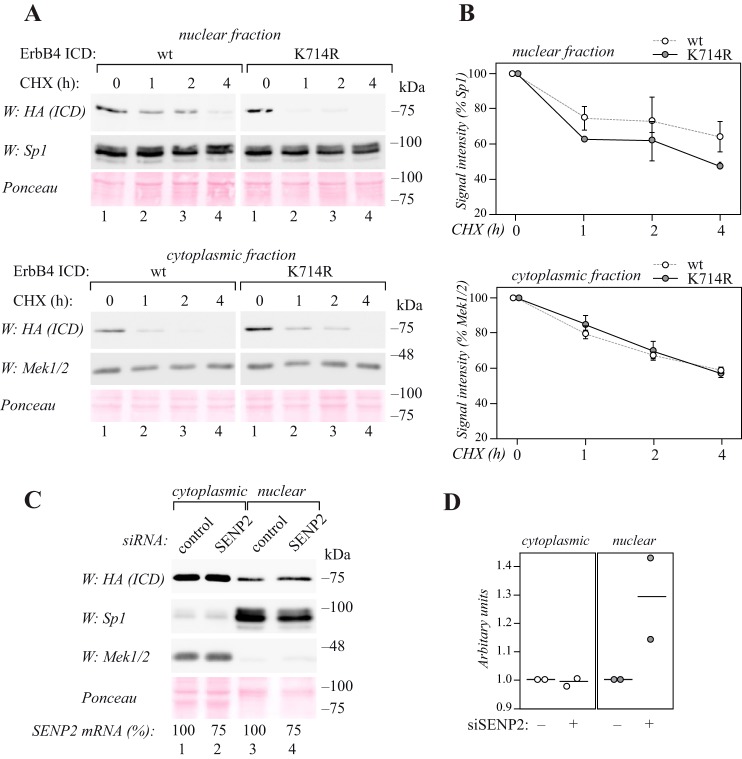
Figure 3.
SUMOylation regulates the nuclear accumulation of ErbB4 ICD. A, MCF-7 cells were transfected with HA-tagged wild-type or K714R ErbB4 ICD, treated with 100 μg/ml cycloheximide (CHX) for the indicated periods of time, and subjected to subcellular fractionation. Nuclear and cytoplasmic fractions were analyzed by Western blotting with anti-HA (H3663, Sigma-Aldrich). Equal loading of the nuclear fraction was controlled with anti-Sp1, and loading of the cytoplasmic fraction with anti-Mek1/2. Total protein loading in both fractions was controlled with Ponceau staining. Representative data of two independent experiments are shown. B, quantification of HA (ErbB4 ICD) signal intensities (shown in A) normalized to Sp1 or Mek1/2. Data are represented as mean ± range from two independent experiments. C, MCF-7 cells were transfected with HA-tagged ErbB4 ICD together with control or SENP2 siRNA and subjected to subcellular fractionation. Nuclear and cytoplasmic fractions were analyzed by Western blotting with anti-HA (H3663, Sigma-Aldrich). Fractionation was controlled with anti-Sp1 and anti-Mek1/2, and total protein loading with Ponceau staining. Efficiency of RNA interference analyzed by qRT-PCR is indicated as relative SENP2 mRNA levels in cells treated with SENP2-targeting siRNA compared with cells treated with control siRNA (SENP2 mRNA %). Representative data of two independent experiments are shown. D, quantification of HA (ErbB4 ICD) signal intensities (shown in C) normalized to Ponceau. The circles indicate individual data points from two independent experiments and the horizontal line indicates the mean.