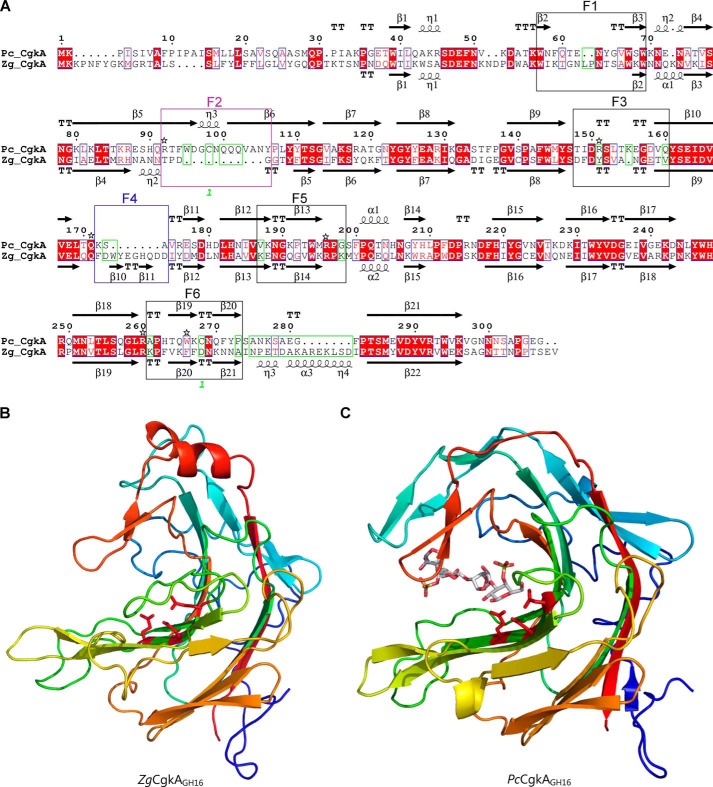
Figure 2.
Comparison of the catalytic modules of PcCgkAGH16 and ZgCgkAGH16. A, multiple-sequence alignment of the catalytic modules of both enzymes. The alignment was produced with MultAlin, and the figure was produced using ESPript version 3.x software. Secondary structure elements are symbolized with arrows for β-sheets, curls for α-helix, and T for turns; the “finger” regions surrounding the catalytic channel are boxed and numbered from F1 to F6; the sequence divergences that have structural consequences are boxed in green; the numbers 1 in green refer to the cysteine residues involved in the single disulfide bond of PcCgkAGH16; the black stars above the sequence highlight the amino acids that were mutated into alanine for PcCgkAGH16 in this study. B, schematic representation of the structural fold of ZgCgkAGH16. The color code is graduated from blue (N terminus) to red (C terminus) along the polypeptide chain. Catalytic residues Glu159-Asp161-Glu164 are shown as sticks and colored in red. C, schematic representation of the structural fold of PcCgkAGH16-E168D in complex with a κ-neocarratetraose, shown in stick representation (carbon atoms are white, oxygen atoms are red, and sulfur atoms are orange). The color code of the polypeptide chain is the same as in B. Catalytic residues Glu163-Asp165-E168D are shown as sticks and colored in red.