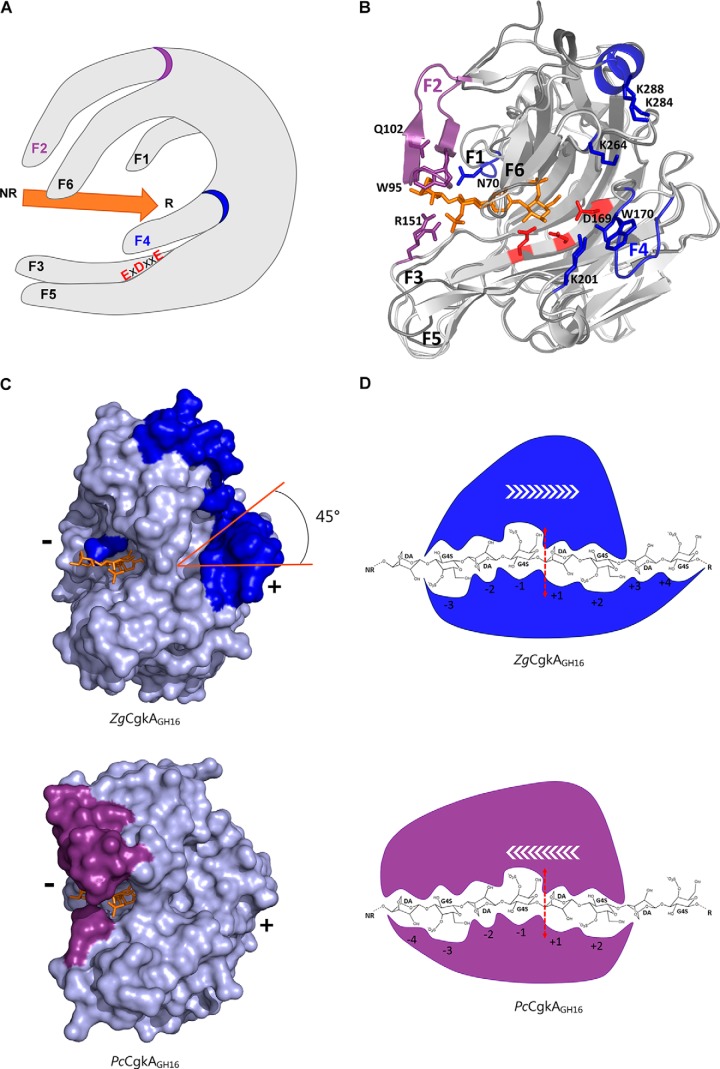
Figure 6.
Structural comparison of the catalytic modules of PcCgkAGH16-E168D and ZgCgkAGH16 and their interactions with κ-neocarratetraose. A, schematic representation of the overall structure with the six identified fingers numbered from F1 to F6; the catalytic triad is identified with red letters; the substrate is symbolized by the orange arrow oriented from its non-reducing (NR) to its reducing (R) end; the specific fingers F2 and F4 are identified with a purple and a blue ring, respectively. B, ribbon representation of the two superposed structures with the oligosaccharide in orange, the catalytic triad in red, and the specific structural features of both enzymes in purple for PcCgkAGH16-E168D and blue for ZgCgkAGH16. C, ZgCgkAGH16 surface representation with its specific structural features in blue (top) superimposed with the oligosaccharide trapped in PcCgkAGH16-E168D in orange and PcCgkAGH16-E168D surface representation with its specific structural features in purple (bottom) and the oligosaccharide in orange. The tilt of the positive substrate binding sites by 45° is highlighted in orange. (+) and (−) indicate the regions of the respective subsites. D, schematic representation of the interactions between the catalytic subsites of ZgCgkAGH16 (blue) and PcCgkAGH16-E168D (purple) with a DP8 κ-carrageenan; the red dashed arrow symbolizes the cleavage site; white arrows symbolize the proposed direction of enzymatic processivity.