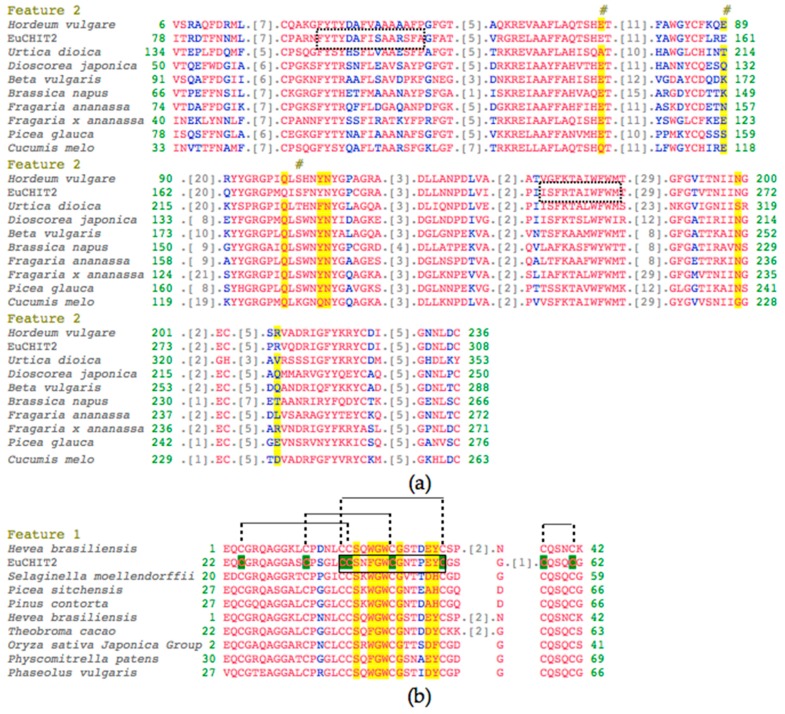
Figure 1.
Alignment analysis of EuCHIT2 with other organisms. (a) Alignment of deduced amino acid sequence of EuCHIT2 with family 19 chitinases different organisms. The two chitinase family 19 signature sequences FYTYDAFISAARSFAGFA (95–117 aa) and ISFRTAIWFWM (221–231 aa) were surrounded with imaginary frames; (b) alignment of deduced amino acid sequence of EuCHIT2 with ChtBD_CH19_hevein (a hevein protein) of other organisms. The chitin recognition domain signature sequence CCSNFGWCGNTPEYC was surrounded with black frame and the eight “C” residues highlighted in green are the conserved cysteine residues that can form four disulfide bonds in the chitin-binding region of EuCHIT2. The species shown in the figure are: 2BAA (Hordeum vulgare), P11218.3 (Urtica dioica), P80052.2 (Dioscorea japonica), and P42820.1 (Beta vulgaris); and Q43391 (Brassica napus), AAD28733.1 (Triticum aestivum), AAF00131.1 (Fragaria x ananassa), Q40838 (Picea glauca), and AAF64475.1 (Cucumis melo); and 1WKX_A (Hevea brasiliensis), EFJ22468.1 (Selaginella moellendorffii), ABK24218.1 (Picea sitchensis), AEF59005.1 (Pinus contorta), 1Q9B_A (Hevea brasiliensis), Q41596.1 (Theobroma cacao), 2DKV_A (Oryza sativa Japonica Group), XP_001767856.1 (Physcomitrella patens), and CAB97002.1 (Phaseolus vulgaris). The red and blue letters are residues in the chitinase domain share high and relatively higher similarity, respectively. The grey number represents the number of amino acid residues that have been omitted. The amino acid residues highlighted in yellow are sugar-binding residues, and the residues under the # markers are catalytic residues.