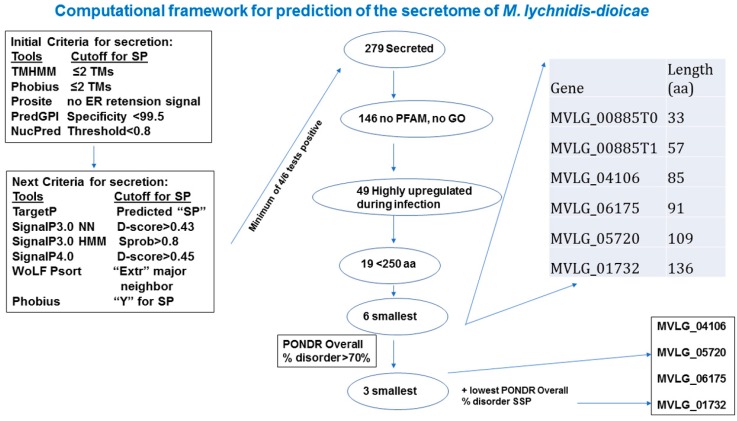
Figure 5.
Computational framework for prediction of secretome for M. lychnidis-dioicae and selection of candidate effectors for further analyses. Detailed description of tools and cut-off criteria for secretome prediction and prediction of disorder are provided in Supplementary Methods. Numbers tally for proteins at each stage of secretome prediction are provided in tab 3 of Table S1. TM, trans-membrane domain; ER, endoplasmic reticulum’ SP, secreted protein; PFAM, protein family; aa, amino acid; GO, gene ontology; MVLG designations refer to specific Microbotryum lychnidis-dioicae proteins.