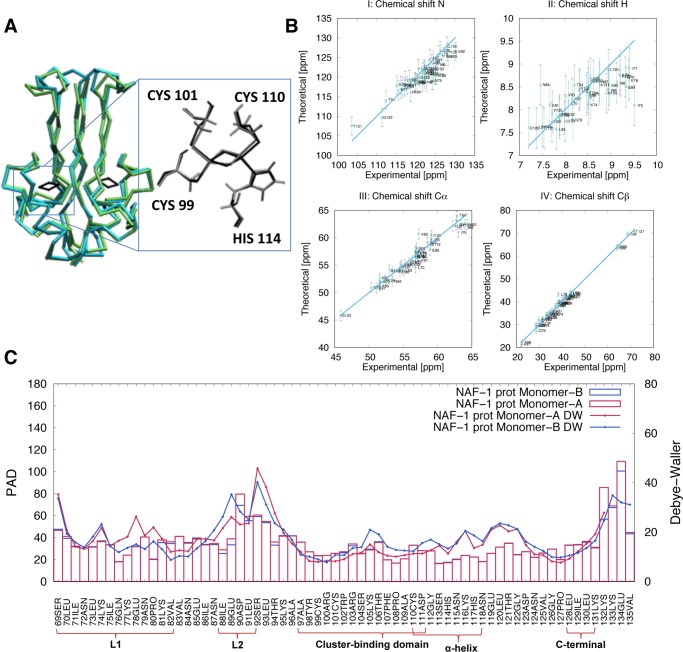
Figure 1.
Structural and flexibility determinants of the NAF-1 protein in the His114:Nε protonated state. (A) Superposition of the main representative MD structure (cyan), as obtained by clusterization (see Section 2.3), with the corresponding X-ray structure (green, pdbID:4OO7). (B) Calculated N, H, Cα, and Cβ chemical shifts for monomers A (violet) and B (green) and corresponding experimental values. The differences were lower than the uncertainties associated with the root mean square error (error bars) of the program used for the chemical shift prediction (SHIFTX249). (C) Protein angular dispersion (PAD) values47 overlaid on the experimental Debye–Waller factors.31 Monomers A and B are colored in red and in blue, respectively. Similar results were obtained for the other systems simulated here (see Section S2.2 and Figure S5).