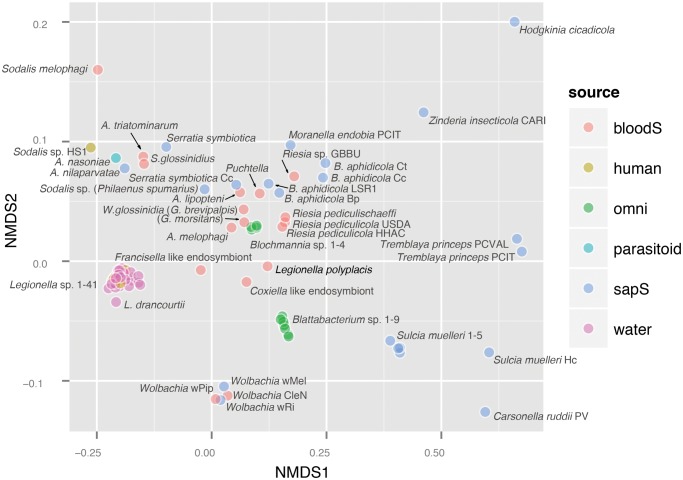
Fig. 4.
—NMDS analysis based on Bray–Curtis dissimilarities calculated for the genome content of selected bacteria analyzed across all COG orthologs. The position of each genome represents as closely as possible the pairwise dissimilarity between the genomes. The legend abbreviations for the bacterial source are as follows: bloodS: blood sucking host, human: clinical isolate, omni: omnivorous insect host, parasitoid: that is, Nasonia vitripennis host, sapS: sap sucking insect host, water: environmental water sample. List of the analyzed genomes is provided in supplementary table S2 in supplementary file S1, Supplementary Material online.