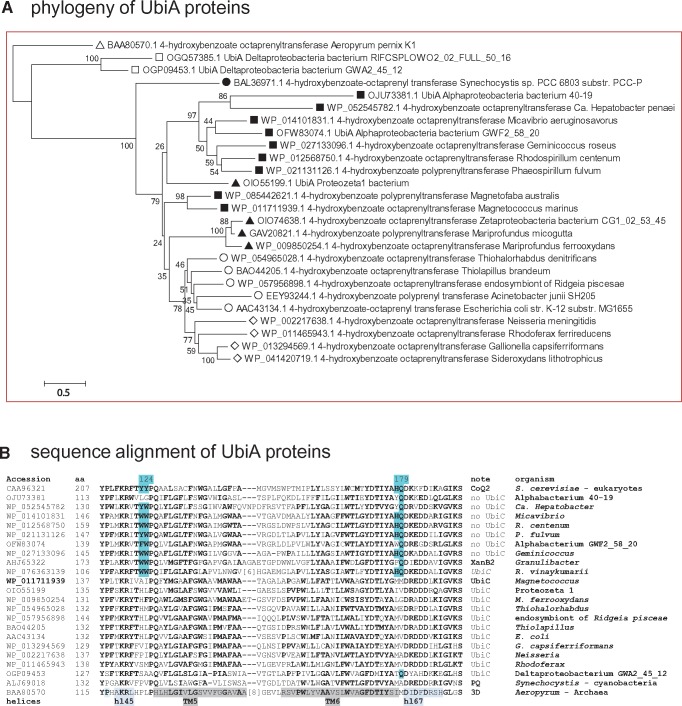
Fig. 2.
—UbiA proteins have different molecular signatures whether they are associated with UbiC or not. (A) Phylogenetic tree of UbiA proteins from various bacterial groups. The NJ tree was obtained after manual refinement of a CLUSTALW alignment of 26 UbiA sequences from phylogenetically diverse taxa using the program MEGA5 (Tamura et al. 2011). The percentage value of 500 bootstraps is shown for each node. After removing all positions containing gaps and missing data, the final data set contained a total of 244 positions. Clearly, the proteins from alphaproteobacteria (black squares) which are not associated with UbiC form a sister group to that including zeta (black triangles), gamma (empty circles), and beta proteobacteria (empty diamonds). Note the precursor position of the cyanobacterial Synechocystis (black circle) and the outgroup position of the Archaean Aeropyrum (white triangle). White squares indicate distant homologous proteins from delta proteobacteria. Similar results were obtained with ML trees. (B) Alignment of conserved regions of UbiA proteins. The same alignment of UbiA proteins used in (A) was reduced to 22 sequences and additionally included the yeast CoQ2 homologue of UbiA. The alignment block shown includes the functionally important regions II and III that were previously deduced (Ohara et al. 2009) and later verified by the 3D structure of UbiA (Cheng and Li 2014). Highlighted in pale blue are the residues conserved among UbiA proteins from taxa without UbiC (indicated as no UbiC) that lie close to the active site at the negative side of the membrane and are structurally different from those of UbiA proteins associated with UbiC, following the numeration of Aeropyrum UbiA (Cheng and Li 2014). The regions corresponding to alpha helices in the 3D structure of Aeropyrum UbiA (Cheng and Li 2014) are highlighted in gray (transmembrane, TM) or light blue.