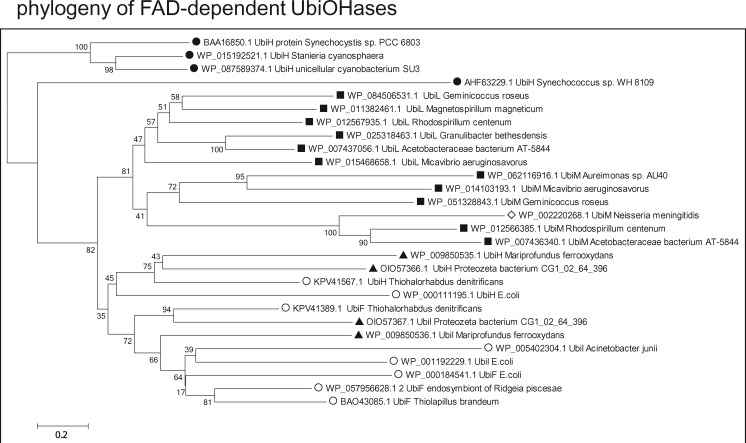
Fig. 4.
—Phylogenetic trees of FAD-dependent Q ring hydroxylases indicate cyanobacterial ancestry. A selection of each variant of FAD-dependent UbiOHases that have been recently categorized (Pelosi et al. 2016) has been chosen to cover the phylogenetic span used to analyze other enzymes involved in Q biosynthesis, for example, UbiA (fig. 2A), whereas matching as much as possible the topology of NJ trees obtained from all the results of wide BLASTP searches, as described earlier (Degli Esposti 2016). The percentage of replicate NJ trees in which the associated taxa clustered together in the bootstrap test is shown next to the branches. The rate variation among sites was modeled with a gamma distribution (shape parameter = 2) and all positions containing gaps were eliminated, for a final data set of 326 amino acid positions. Note how the UbiOHases of proteobacteria clearly originate from UbiH proteins of cyanobacteria (black circles), segregating into two sister groups (cf. Pelosi et al. 2016): the top group contains UbiL and UbiM homologues that are predominantly present in alphaproteobacteria (black squares) and a few betaproteobacteria (empty diamonds), whereas the bottom group includes UbiH and the closely related UbiI and UbiF proteins, which are present in zeta (black triangles) ans gamma proteobacteria (empty circles), but generally absent from alpha proteobacteria. Similar trees showing the same separation of groups of Ubi hydroxylases have been obtained with the ML method and different numbers of bootstrap replicates. The original annotation of most proteins has been modified for providing a homogeneous representation of the various types of FAD-dependent hydroxylases.