Abstract
RAD51, a key factor in homology-directed repair (HDR), has long been considered an attractive target for cancer therapy, but few specific inhibitors have been found. A cell-penetrating, anti-DNA, lupus autoantibody, 3E10, was previously shown to inhibit HDR, sensitize tumors to radiation, and mediate synthetic lethal killing of BRCA2-deficient cancer cells, effects that were initially attributed to its affinity for DNA. However, as the molecular basis for its ability to inhibit DNA repair, we report that 3E10 directly binds to the N-terminus of RAD51, sequesters RAD51 in the cytoplasm, and impedes RAD51 binding to DNA. Further, we generate separation-of-function mutations in the complementarity-determining regions of 3E10 revealing that inhibition of HDR tracks with binding to RAD51 but not to DNA, whereas cell penetration is linked to DNA binding. The consequences of these mutations on putative 3E10 interactions with RAD51 and DNA are correlated with in silico molecular modeling. Taken together, the results identify 3E10 as a novel inhibitor of RAD51 by direct binding, accounting for its ability to suppress HDR and providing the molecular basis to guide pre-clinical development of 3E10 as an anti-cancer agent.
INTRODUCTION
Antibody therapy for cancer provides a powerful tool to specifically target factors that support the malignant phenotype. Currently, more than a dozen antibodies have been approved by the FDA for cancer therapy (1). Many of these antibodies target mutant or overexpressed cell surface receptors such as EGFR or HER2. Additional antibody targeting strategies include binding to surface markers specific to malignant cells or extracellular ligands that promote tumor growth and/or neovascularization of hypoxic tumors (e.g. VEGF) (1–3).
The discovery that inhibitors of poly(ADP) ribose polymerase (PARP) selectively kill cells deficient in homology-directed repair (HDR) has led to a new focus on therapeutic exploitation of DNA repair pathways (4–6). Numerous human malignancies with mutations in HDR genes, such as BRCA1 and BRCA2, have been successfully treated in clinical trials with PARP inhibitors leading to the FDA approval of Olaparib for the treatment of ovarian cancer. DNA repair functions are confined mainly within the nucleus of a cell, and so pharmacological strategies have so far focused on small molecules rather than antibodies since cellular uptake of antibodies poses a formidable obstacle (7).
DNA double-strand breaks (DSBs) are the most deleterious form of DNA damage and are generated by radiation therapy and numerous chemotherapy agents. In mammalian cells, DSBs are repaired by two main pathways: non-homologous end-joining (NHEJ) and homology-directed repair (HDR). During HDR, DSBs are processed by an assembly of nucleases to create 3′ single-stranded DNA (ssDNA) tails (8–10). The resected 3′ ssDNA tails are initially stabilized and bound by replication protein-A (RPA). RPA complexes on the ssDNA are subsequently replaced by RAD51 aided by the actions of mediator proteins such as BRCA2 (11–13). The RAD51 protein forms a helical nucleoprotein filament on the ssDNA facilitating strand invasion and the homology search usually within the sister chromatid (8,14). RAD51 is highly conserved among eukaryotes and is essential for HDR and cell viability (15). Many human cancers express elevated levels of RAD51 (16) leading to chemotherapy and radiation resistance (16–21). Consequently, RAD51 has been considered an attractive target for cancer therapy (15). The Connell group recently identified a promising small molecule inhibitor of RAD51, but increases in potency will be needed for clinical development (15).
Interestingly, a small number of systemic lupus erythematosus autoantibodies have been found to penetrate living cells (22). One such antibody is 3E10, a cell penetrating, anti-DNA antibody that is non-toxic to normal cells and has been investigated as a delivery vehicle for various conjugates, primarily using single chain variable fragments (scFvs) derived from it (23). Cellular penetration by 3E10 has been linked to its ability to bind DNA, as DNA binding mutants of 3E10 are unable to penetrate cells (24). While the exact molecular basis for 3E10 internalization has yet to be determined, it has been shown to depend on the equilibrative nucleotide transporter 2 (ENT2) (25).
Recently, our group discovered that 3E10 treatment of human cells inhibits DNA DSB repair by HDR, confers sensitivity to ionizing radiation, and mediates synthetic lethality in BRCA2-deficient cancer cells (26). Biochemically, we determined that 3E10 reduces the efficiency of RAD51-mediated strand exchange, but we attributed this result to competition for binding sites between 3E10 and RAD51 for the ssDNA substrate (26).
Here, we report the unexpected finding that 3E10 physically interacts with RAD51, defining a new molecular basis for HDR inhibition by 3E10. Utilizing purified 3E10 scFv protein and purified fragments of RAD51, we demonstrate that 3E10 binds to the N-terminal domain of RAD51, a region essential for homo-oligomerization and crucial for RAD51 filament formation. We find that 3E10 inhibits RAD51 accumulation on ssDNA and RAD51-dependent DNA strand exchange. Further in keeping with this mechanism of action, 3E10 inhibits RAD51 foci formation in response to ionizing radiation or etoposide, a measurement of a cell's ability to form RAD51 nucleoprotein filaments at sites of DNA damage. Mutational analysis of the 3E10 variable region reveals separation-of-function linking RAD51 binding to inhibition of HDR and DNA binding to cell penetration. Because we have shown in pre-clinical mouse studies that 3E10 can radiosensitize tumors and suppress the growth of BRCA2-deficient tumors (26), 3E10 has attracted interest for clinical development. By identifying RAD51 as the functional target of 3E10, our results serve to better define a clinical development path for 3E10 and provide the basis for further molecular optimization by affinity maturation.
MATERIALS AND METHODS
Cloning of 3E10 scFv and site directed mutagenesis
The single chain variable fragment of 3E10 was cloned into a phCMV1_2XMBP expression vector to aid in purification. Point mutations were introduced into the heavy or light chain regions of the scFv using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). The plasmid was sequenced to ensure no undesired mutations were introduced during PCR.
Purification of 2XMBP-scFvs
The 2XMBP expression constructs were transfected into suspension Expi293F Cells (Life Technologies) with polyethylenimine (PEI linear, MW ∼25 000 from Polysciences, Inc.). Briefly, the plasmid DNA was diluted in OptiPro SFM (Gibco), mixed with PEI, and added to Expi293F cells in shake flasks. Sixteen to eighteen hours later, BalanCD CHO FEED 3 (Irvine Scientific) and Valproic acid sodium salt (Sigma-Aldrich, 3.8 mM final concentration) were added to enhance expression. The cells were grown at 37°C and 8% CO2 for three additional days. The cells were harvested by centrifugation. All successive steps in the purification were performed at 4°C. The cell pellet was resuspended in lysis buffer (50 mM HEPES pH 7.5, 250 mM NaCl, 1% NP-40, 1 mM MgCl2, 1× Protease Inhibitor Cocktail (Roche complete, EDTA-free), 1 mM DTT) and rocked at 4°C for 15 min. The lysate was cleared by centrifugation at 7000 rpm for 15 min. The cleared lysate was incubated with amylose resin (New England BioLabs, Inc.) for 2 h. The lysate containing the amylose bead matrix was spun down and the supernatant was removed. The amylose beads were washed three times with wash buffer (50 mM HEPES pH 7.5, 250 mM NaCl, 0.5 mM EDTA, 1 mM DTT). The bound protein was eluted from the amylose beads in wash buffer containing 10 mM maltose. The eluate containing the 2xMBP-scFv was buffer exchanged into PBS and concentrated.
Electrophoretic mobility shift assay
The DNA binding ability of each of the 2XMBP-scFvs was determined by an electrophoretic mobility shift assay. Increasing amounts of the purified 2XMBP-scFv proteins (1, 2 or 5 μM) were added to 10 nM radiolabeled 167mer ssDNA (32P-radiolabeled) in Buffer S (25mM TrisOAc (pH 7.5), 1 mM MgCl2, 2 mM CaCl2, 0.1 μg/μl BSA, 2 mM ATP and 1 mM DTT). The reactions were incubated for 30 min at 4°C. The products were separated using 6% polyacrylamide gel electrophoresis and analyzed using a phosphorimager. Experiments were performed at least three times for each 2XMBP-scFv protein.
Immunofluorescence—cell penetration of purified 2xMBP-scFv
U2OS cells were pretreated with purified 2XMBP-scFv proteins for 24 hours in chamber well slides (Millipore Millicell EZ Slides). Cells were fixed with 1% paraformaldehyde/2% sucrose for 15 minutes at room temperature, followed by 100% methanol for 30 min at –20°C, and 50% methanol/50% acetone for 20 min at –20°C. Slides were then incubated in permeabilization/blocking solution (10% BGS, 0.5% Triton X-100 in phosphate-buffered saline (PBS)) at room temperature for 1 h. Primary antibody (mouse anti-MBP monoclonal antibody, New England BioLabs, Inc. #E8032S) was diluted 1:500 in permeabilization/blocking solution and used to stain cells at 4°C overnight. The secondary antibody used was Alexa Fluor 594-conjugated goat anti-mouse immunoglobulin G (IgG) (Life Technologies). Cells were costained with DAPI to visualize the nuclei. Slides were imaged on a Zeiss-CARV II confocal microscope.
Immunofluorescence-transfected 2xMBP-scFv expression constructs
Primary skin human fibroblasts were seeded in chamber well slides (Millipore Millicell EZ Slides) and were left untransfected, transfected with the truncated 2XMBP-scFv expression construct, or transfected with one of the four 2XMBP-scFv expression constructs. Cells were fixed and stained as described above each day for up to 3 days post-transfection.
HDR host cell reactivation assays
Luciferase reporter DNA plasmids for HDR were digested with I-SceI (New England Biolabs, Inc. #R0694) according to I-SceI manufacturer instructions. Digestion was confirmed on a 0.7% agarose gel and then purified using the QIAquick PCR Purification Kit (QIAGEN # 28104). Primary human skin fibroblasts were seeded at 1 × 105 cells per well in a 12-well dish the day before transfection. For experimental conditions, 4 μg of the digested plasmid and 50 ng of a Ranilla plasmid were transfected with Lipofectamine 3000 Reagent (Thermo Fisher Scientific) according to manufacturer instructions. For control conditions, 2 μg of a control plasmid and 50 ng of a Renilla plasmid were transfected with Lipofectamine 3000 Reagent (Thermo Fisher Scientific) according to manufacturer instructions. Cells were collected and lysed in 1× Passive Lysis Buffer (Promega) 48 hours after transfection. Lysates were run in triplicate in a 96-well plate and analyzed using the Dual-Luciferase® Reporter Assay System (Promega) and a Synergy HT plate reader (BIO-TEK). The percent reactivation was calculated. Experiments were performed at least three times for biological replicates. Each experiment contained technical triplicates per condition. Statistical significance was determined using the unpaired t-test (GraphPad Prism).
NHEJ host cell reactivation assays
U2OS EJ5 reporter cell lines were seeded in triplicate for each condition and were left untreated, pre-treated media control, or pre-treated with 10μM purified full length 3E10. The next day, the I-SceI expression plasmid (4 μg) was transfected into 1 × 106 cells per replicate using the Amaxa Nucleofector (Lonza) 72 hours before analysis. Cells were analyzed for GFP expression by flow cytometry, and data were analyzed using the FlowJo software (Tree Star Inc.). Luciferase reporter DNA plasmids for NHEJ were digested with HindIII (New England Biolabs, Inc. #R3104) according to HindIII manufacturer instructions. Digestion was confirmed on a 0.7% agarose gel and then purified using the QIAquick PCR Purification Kit (QIAGEN # 28104). Primary human skin fibroblasts were seeded at 1 × 105 cells per well in a 12-well dish the day before transfection. Transfection steps were carried out as above. Cells were collected and lysed in 1× Passive Lysis Buffer (Promega) 24 h after transfection and analysis was continued as above. Each experiment contained technical triplicates per condition. Statistical significance was determined using the unpaired t-test (GraphPad Prism).
Affinity pull-downs from 293T cells transfected with D31N 2XMBP-scFv and 2XMBP-BRC1-8 region of BRCA2
Human 293T cells (5 × 105 cells/well) were transiently transfected in 6-well plates with 1 μg of the indicated construct using TurboFect transfection reagent (ThermoFisher Scientific). Cellular lysates were harvested 36 h post-transfection using buffer ‘B’ (50 mM HEPES (pH 7.5), 250 mM NaCl, 1% NP-40, 1 mM MgCl2, 1 mM DTT, 250 units/ml Benzonase (EMD Millipore) and 1× cOmplete EDTA-free Protease Inhibitor Cocktail (Roche)). Cell lysates were batch bound to amylose resin for one hour, washed 3× in wash buffer (50 mM HEPES (pH 7.5), 250 mM NaCl, 0.5 mM EDTA), and then incubated with 1 μg of purified human RAD51 for 30 min at 37°C. The bead complexes were then washed again 3× with wash buffer including 1% NP-40 and finally eluted with 10 mM maltose. Elutions were run on 4–15% gradient SDS-PAGE gels (Bio-Rad StainFree TGX). The proteins were visualized by SyproOrange (Invitrogen) staining and scanning on a Storm 860 PhosphorImager (Molecular Dynamics).
Affinity western from hela cells transfected with full length 3E10 expression constructs
HeLa cells were left untransfected or transfected with expression full length 3E10 expression constructs. Three days later, cells were lysed in AZ lysis buffer (50 mM Tris pH 8, 250 mM NaCl, 1% NP-40, 0.1% SDS, 5 mM EDTA, 10 mM Na4P2O7, 10 mM NaF, 1× cOmplete EDTA-free Protease Inhibitor Cocktail (Roche), 1× PhosSTOP (Roche)), either in the presence or absence of benzonase, and the protein concentration of each sample was determined using the DC™ (detergent compatible) protein assay (Bio-Rad Laboratories, Inc.). Protein A/G bead slurry (New England Biolabs, Inc.) was prewashed in wash buffer (50 mM HEPES pH 7.5, 250 mM NaCl, 0.5 mM EDTA) and then 500 μg of each lysate was incubated with the protein A/G beads overnight at 4°C. The next day, the supernatant was removed, the beads were washed three times in wash buffer, and the immobilized proteins were eluted in 5× Laemmli sample buffer at 90°C for 5 min. Samples were run on a 7.5% gel and transferred for western blot.
The primary antibodies used were rabbit anti-RAD51 (H-92 Santa Cruz Biotechnology #sc-8349), rabbit anti-RAD52 (Cell Signaling Technology #3425), rabbit anti-BRCA1 (C-20 Santa Cruz Biotechnology #sc-642), mouse anti-KU80 (BD Biosciences #611360), rabbit anti-γH2Ax Ser 139 (20E3 Cell Signaling Technology #9718), rabbit anti-MRE11 (H-300 Santa Cruz Biotechnology #sc-22767), mouse anti-CHK1 (2G1D5 Cell Signaling Technology #2360), rabbit anti-RAD50 (Cell Signaling Technology #3427), rabbit anti-BRCA2 (Cell Signaling Technology #9012), rabbit anti-PTEN (D4.3 Cell Signaling Technology #9188), rabbit anti-AKT1 (C73H10 Cell Signaling Technology #2938), rabbit anti-NBS1/p95 (D6J5I Cell Signaling Technology #14956), rabbit anti-ATRIP (Cell Signaling Technology #2737), rabbit anti-53BP1 (Cell Signaling Technology #4937), rabbit anti-XRCC3 (Thermo Scientific PA1-16535), rabbit anti-RPA70/RPA1 (Cell Signaling Technology #2267), mouse anti-Phospho RPA (S4/S8) (Bethyl Laboratories, Inc. #A300-245A) and rabbit anti-PALB2 (Sigma Aldrich # SAB3500079).
Primary antibodies were used at 1:500 dilutions and were incubated for 2–3 h at room temperature or overnight at 4°C. Secondary goat-anti-rabbit or goat anti-mouse antibodies (Thermo Fisher Scientific/Pierce) or were used at a 1:10 000 dilution for 1 h at room temperature. Primary and secondary antibodies were prepared in 5% milk. Three washes with Tris Buffered Saline with Tween 20 were each performed after primary incubation and after secondary incubation. Membranes were developed using SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific).
Affinity western from 293T cells transfected with 2XMBP-scFv expression constructs
293T cells (5 × 105 cells/well) were transiently transfected in 6-well plates with 1 μg of the indicated construct using TurboFect transfection reagent (ThermoFisher Scientific). Cellular lysates were harvested 36 h post-transfection using buffer ‘B’ (50 mM HEPES (pH 7.5), 250 mM NaCl, 1% NP-40, 1 mM MgCl2, 1 mM DTT, 250 units/ml Benzonase (EMD Millipore) and 1× cOmplete EDTA-free Protease Inhibitor Cocktail (Roche)). Cell lysates were batch bound to amylose resin overnight, washed 3× in wash buffer (50 mM HEPES (pH 7.5), 250 mM NaCl, 0.5 mM EDTA), and then eluted in 6× Laemmli sample buffer at 90°C for 5 min. Elutions were run on 4–15% gradient SDS-PAGE gels and then transferred for western blot. Western blots were continued as described above. The primary antibodies used were rabbit anti-RAD51 (H-92 Santa Cruz Biotechnology #sc-8349) and rabbit anti-MBP probe (C-18 Santa Cruz Biotechnology #sc-808). The experiment was repeated four times and the relative levels of RAD51 bound to the expressed 2XMBP-scFv was plotted (combined levels from four experiments; normalized to MBP levels and then normalized to WT 2XMBP-scFv). Statistical significance was determined using the unpaired t-test (GraphPad Prism).
Affinity binding assay and Kd calculations
Purified WT and D31N scFv proteins at a fixed concentration (400 ng; 3.64 pmol; 72.7 nM final concentration) were incubated with increasing amounts of purified RAD51(100, 200, 800 and 1600 ng; 2.7, 5.41, 21.62 and 43.24 pmol; and 54, 108, 432 and 864 nM, respectively) in a total volume of 50 μl for 30 min at 37°C. Amylose resin and binding buffer [50 mM HEPES (pH 7.5), 250 mM NaCl, 0.5 mM EDTA and 1 mM DTT] were added and the reaction was gently rocked for 1 h at 4°C. The bound complex was then washed with binding buffer and 0.1% Igepal CA-630. The proteins were eluted from the amylose beads with 6× Laemmli sample buffer and the samples were heated at 54°C for 4 min. Samples were run on a 4–15% gradient SDS-PAGE gel. The proteins were visualized by staining with SyproOrange (Invitrogen). The protein bands were imaged on a Storm 860 PhosphorImager (Molecular Dynamics) and quantified using ImageJ. The amount of RAD51 pulled down with 2XMBP-scFv proteins was determined using standard curves generated from known concentrations of RAD51 and 2XMBP-scFv proteins run in parallel. The Kd values were calculated in GraphPad Prism using a hyperbolic standard curve interpolation. Experiments were performed at least three times for biological replicates.
Purification of human RAD51 fragments
pGEX-6P-1 GST expression constructs (GE Healthcare) containing the human RAD51 fragments were a gift of Patrick Sung. Briefly, the RAD51 constructs were expressed in the Rosetta (Novagen) derivative strain of BL21 Escherichia coli cells. The cells were grown at 37°C until the OD reached 0.6 and then induced with IPTG at 25°C for 4 h. The cells were harvested by centrifugation at 5000 rpm for 10 min at 4°C. All successive steps in the purification were performed at 4°C. The cell pellet was resuspended in breaking buffer (50 mM Tris–HCl pH 7.5, 1 mM EDTA, 10% sucrose, 250 mM KCl, 1 mM β-mercaptoethanol, 2 mM DTT, 0.01% Igepal, 0.1 mg/ml lysozyme, 5 mM benzamide, 1 mM PMSF and a combination of protease inhibitors at 5 μg/ml: aprotinin, chymostatin, leupeptin and pepstatin A) followed by sonication. The lysate then underwent ultracentrifugation at 40 000 rpm for 1 h. The cleared lysate was incubated with Glutathione Sepharose (GE Healthcare) for 90 min. The lysate containing the Glutathione matrix was poured into a glass column (BioRad). After three washes with wash buffer (20mM KH2PO4 pH 7.5, 10% glycerol, 1 mM EDTA, 1 mM DTT, 150 mM KCl), the bound protein was eluted from the glutathione beads in wash buffer containing reduced glutathione. The eluate containing the RAD51 fragments was loaded onto a primed gel filtration column. The samples were eluted off the column in 1 M tris buffer containing 20 mM glutathione. The peak fractions containing RAD51 were pooled, concentrated, and frozen in small aliquots at –80°C.
Protein pull-downs
2 μM purified 2XMBP-3E10 scFvs were immobilized on amylose bead slurry (New England Biolabs) prewashed in washed in wash buffer (50 mM HEPES pH 7.5, 250 mM NaCl, 0.5 mM EDTA) for 30 min at 4°C in Buffer S (25 mM TrisOAc pH 7.5, 1 mM MgCl2, 2 mM CaCl2, 0.1 μg/μl BSA, 2mM ATP and 1mM DTT). 2.5 μM purified full length RAD51 or GST-tagged RAD51 fragment was added and the reaction continued for another 30 min at 4°C. The supernatant was removed, and the beads were washed three times with wash buffer. The immobilized proteins were eluted in 6× Laemmli sample buffer at 90°C for 5 min. Samples of the supernatant, wash, and eluate were separated on a 7.5% gel and transferred for western blot. Western blots were continued as described above. The primary antibodies used were rabbit anti-RAD51 (H-92 Santa Cruz Biotechnology #sc-8349), rabbit anti-GST (91G1 HRP conjugate Cell Signaling Technology #5475), and rabbit anti-MBP probe (C-18 Santa Cruz Biotechnology #sc-808). A protein pull-down was also performed using 2 μM purified D31N 2XMBP-3E10 scFv and 2.5 μM purified RAD52. This assay was performed as above. Samples of the supernatant, wash, and eluate were separated on a 7.5% gel and the gel was stained with coomassie and imaged using a BioRad ChemiDoc XRS+ imager.
RAD51 DNA binding assays
240 nM of RAD51 and either 1 or 7.5 μM of the purified 2XMBP-scFv proteins were incubated for 5 min at 37°C in Buffer S. Final volumes were normalized with Buffer S as needed. Then, 1 nM of biotinylated 167-mer ssDNA was added to the reaction and the samples were incubated for an additional 5 min at 37°C. The DNA-protein complexes were then captured by adding 2.5 μl of pre-washed MagnaLink Streptavidin magnetic beads (SoluLink) in buffer SW (Buffer S supplemented with 0.1% Igepal CA-630 and 0.5% Triton X-100). The bead–DNA–protein complexes were incubated with agitation at 25°C for 5 min. The supernatant was removed and the beads were washed three times with buffer SW’ (Buffer SW lacking 2 mM ATP). The bead samples were resuspended in 20 μl 6× Laemmli sample buffer, heated at 54°C for 4 min, and loaded onto a 7.5% gradient SDS-PAGE gel. The RAD51 proteins bound and eluted from the biotinylated DNA substrate were detected by western blot as above. The percent of RAD51 bound to DNA was calculated using ImageJ. Experiments were performed at least three times for biological replicates. Statistical significance was determined using the unpaired t-test (GraphPad Prism).
Strand exchange assays
An optimal concentration of RAD51 for maximum percent strand exchange was first determined using a titration of RAD51 (from 0.1 to 0.45 μM, with 0.22 μM RAD51 being the optimal concentration). The strand exchange reactions were then performed with either 1 or 7.5 μM of the purified 2XMBP-scFv proteins. Experiments were performed at least three times for biological replicates. Statistical significance was determined using the unpaired t-test (GraphPad Prism). 0.22 μM of RAD51 and either 1 or 7.5 μM of the purified 2XMBP-scFv proteins were incubated for 5 min at 37°C in strand exchange buffer (1 M Tris-OAc pH 7.5, 100 mM MgCl2, 100 mM CaCl2, 1 ug/ul BSA, 180 mM ATP, 100 mM DTT). Final volumes were normalized with Buffer S as needed. Then, 100 nM 3′Tail DNA was added to the reaction and the samples were incubated for an additional 5 min at 37°C. Radiolabeled duplex DNA (100 nM 32P-radiolabeled) was added to the reactions followed by a 30 min incubation at 37°C before being deproteinized with the addition of SDS (0.5%) and Proteinase K (0.5 μg/ml) for 10 min at 37°C. The reaction products were separated using 6% native polyacrylamide gel electrophoresis, and analyzed using a phosphorimager. The percent strand exchange was determined for each condition using ImageJ.
Immunofluorescence—RAD51 foci formation and cytoplasmic fraction after IR
Human primary skin fibroblast cells were pretreated with purified 2XMBP-scFv for 24 h in chamber well slides (Millipore Millicell EZ Slides), or transfected with 2XMBP-scFv expression constructs. Cells were then irradiated with 10Gy IR on an X-RAD 320 X-ray irradiation system. Cells were fixed and stained as described above. The primary antibody used was rabbit anti-RAD51 (H-92 Santa Cruz Biotechnology #sc-8349). The secondary antibody used was Alexa Fluor 488-conjugated goat anti-rabbit immunoglobulin G (IgG) (Life Technologies). Cells were costained with DAPI to visualize the nuclei and anti-MBP to visualize the 2XMBP-scFv. Slides were imaged on a Zeiss-CARV II confocal microscope. Images were visualized and quantified using ImageJ. Experiments were performed at least three times for biological replicates. Statistical significance was determined using the unpaired t-test (GraphPad Prism). Human primary skin fibroblast cells were also pretreated with purified full length 3E10 for 24 h in chamber well slides (Millipore Millicell EZ Slides). Cells were then irradiated with 10 Gy IR on an X-RAD 320 X-ray irradiation system. Cells were fixed and stained as described above. The primary antibody used was rabbit anti-RAD51 (H-92 Santa Cruz Biotechnology #sc-8349). The secondary antibody used was Alexa Fluor 488-conjugated goat anti-rabbit immunoglobulin G (IgG) (Life Technologies). Cells were costained with DAPI to visualize the nuclei. Slides were imaged on a Zeiss-CARV II confocal microscope. Images were visualized and quantified using ImageJ. Experiments were performed at least three times for biological replicates. Statistical significance was determined using the unpaired t-test (GraphPad Prism).
Immunofluorescence—pRPA foci formation after IR
Human primary skin fibroblasts cells were pretreated with purified 2XMBP-scFv for 24 h in chamber well slides (Millipore Millicell EZ Slides), or transfected with 2XMBP-scFv expression constructs. Cells were then irradiated with 10 Gy IR on an X-RAD 320 X-ray irradiation system. The cytoplasm was removed according to the Fabrizio d’Adda di Fagagna group protocol. Briefly, cells were incubated with cytoskeleton buffer (10 mM Pipes pH 6.8, 100 mM NaCl, 30 mM sucrose, 3 mM MgCl2, 1 mM EGTA, 0.5% Triton-X 100) for 5 min on ice. Cells were then incubated with cytoskeleton stripping buffer (10 mM Tris–HCl pH 7.4, 10 mM NaCl, 3 mM MgCl2, 1% Tween 40, 0.5% sodium deoxycholate) for 5 min on ice. The cells were then washed three times with cold PBS. The cells were then fixed, stained and imaged as described above. The primary antibody used was mouse anti-Phospho RPA (S4/S8) (Bethyl Laboratories, Inc. #A300-245A). The secondary antibody used was Alexa Fluor 488-conjugated goat anti-mouse immunoglobulin G (IgG) (Life Technologies). Cells were costained with DAPI to visualize the nuclei. Experiments were performed at least three times for biological replicates. Statistical significance was determined using the unpaired t-test (GraphPad Prism).
Immunofluorescence—RAD51 foci formation and cytoplasmic fraction after etoposide
Human primary skin fibroblast cells were pretreated with purified 2XMBP-scFv for 24 h in chamber well slides (Millipore Millicell EZ Slides). The media was exchanged and cells were then treated with 5 μM etoposide (Sigma-Aldrich E1383) in DMSO. Cells were fixed, stained, and imaged as described above. The primary antibody used was rabbit anti-RAD51 (H-92 Santa Cruz Biotechnology #sc-8349). The secondary antibody used was Alexa Fluor 488-conjugated goat anti-rabbit immunoglobulin G (IgG) (Life Technologies). Cells were costained with DAPI to visualize the nuclei and anti-MBP to visualize the 2XMBP-scFv. Experiments were performed at least three times for biological replicates. Statistical significance was determined using the unpaired t-test (GraphPad Prism).
Clonogenic survival assays in PEO-1 and U251 cells
PEO-1 and U251 cells were transfected with 1μg 2XMBP-scFv expression constructs. Two days later, transfected cells were reseeded in triplicate into 6-well plates at low density into Etoposide. Non-transfected cells were also reseeded as a control. Media was replaced 24 hours later. Cells were cultured for 1 to 2 weeks until colonies had formed. Colonies on the 6-well plates were permeabilized with 0.9% saline solution and then stained with crystal violet in 80% methanol for visualization. Colonies of ≥50 cells were quantified. U251s were also left untreated or were pre-treated with purified 2XMBP-scFv proteins for 24 h. Cells were reseeded as above into Etoposide. The experiment was continued as above.
Structural modeling
A predicted structure of 3E10 scFv was generated using SWISS-MODEL. An electrostatic map was generated in the PyMOL Molecular Graphics System (Version 1.8 Schrödinger, LLC.). The scFv-RAD51 interaction model was generated using the Vakser Lab Protein-Protein Docking Web Server from the Center for Computational Biology at the The University of Kansas. (38,39).
RESULTS
3E10 scFv proteins exhibit different biochemical properties
Previous work demonstrated that both the full-length 3E10 antibody and the single chain variable fragment (scFv) could penetrate cells and mediate effects on DNA repair (26). In order to minimize the risk of non-specific antibody-dependent cell-mediated cytotoxicity (ADCC) via the Fc region of 3E10, we have elected to focus on the scFv rather than the full length antibody as a basis for clinical development. This scFv contains the variable region of the heavy chain connected to the variable region of the light chain by a flexible linker, as schematized in Figure 1A.
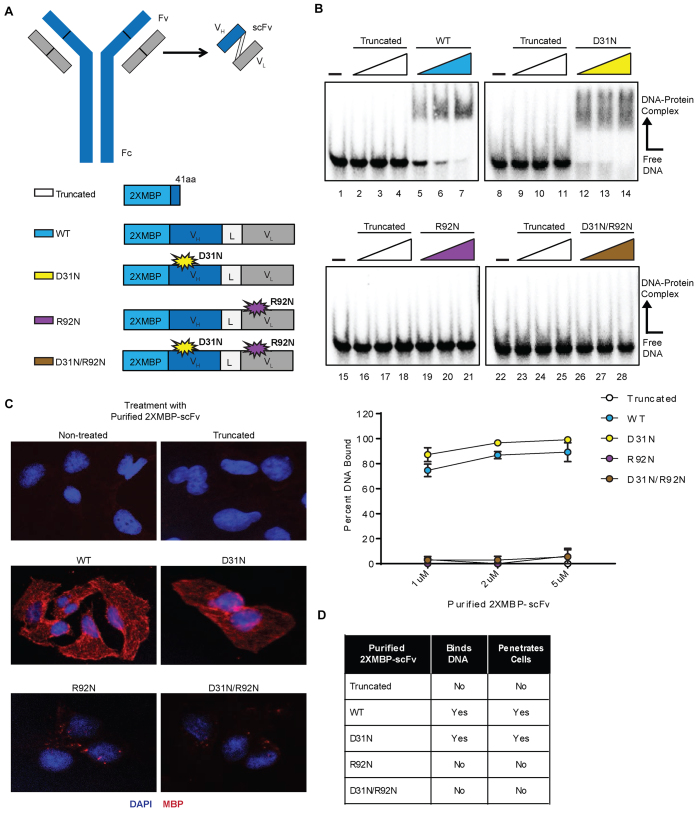
Figure 1.
Purification and biochemical characterization of 3E10-scFv proteins supports the link between 3E10’s DNA binding ability and capacity for cellular penetration. (A) Schematic of the 3E10 full length antibody as well as the derived scFv. The heavy chain (VH) and light chain (VL) are connected by a flexible linker (L) to produce the scFv. The scFv was cloned into a 2XMBP_phCMV1 expression vector. Various point mutations were made to produce five total 2XMBP-scFv proteins. Expression constructs for each scFv and their respective point mutations are schematized. (B) An electrophoretic mobility shift assay was performed using radiolabeled ssDNA to determine the DNA binding ability of all five purified proteins. The assay was performed using a titration of 1, 2 and 5 μM of each purified scFv. Representative images and quantification are shown. (C) U2OS cells were treated with the five scFv proteins and the cellular penetration ability of each was observed via immunofluorescence. (D) A summary of the biochemical properties of each of the five purified scFvs is provided.
To further probe the properties of 3E10 and to conduct mutational analyses, we cloned the 3E10 scFv cDNA into a mammalian expression vector (phCMV1) containing two tandem repeats of the maltose binding protein tag (2XMBP) at the N-terminus of scFv. The MBP tags serve to facilitate affinity purification as well as to enhance solubility and stability of the recombinant proteins (27). Previous results showed that changing the aspartic acid at residue 31 in CDRl of the 3E10 heavy chain to asparagine (D31N) increased 3E10 scFv's binding to both ssDNA and dsDNA (28). In the presence of this D31N mutation, an additional mutation of the arginine residue at position 92 in CDR3 of the light chain to asparagine (R92N) was found to disrupt binding to DNA (24). We introduced either the heavy chain mutation (D31N) or the light chain mutation (R92N) into separate constructs or the combination of both mutations (D31N/R92N) into a single construct in our 2XMBP-scFv expression vector. A premature stop codon (41 amino acids into the scFv) was also introduced to form a truncated protein as a control. The R92N mutation alone has not been previously investigated. A schematic of the expression constructs for each of the five scFvs studied is also provided in Figure 1A.
The scFv constructs were expressed and affinity purified from human 293 cells. First, we determined the DNA binding ability of all five purified scFv proteins. An electrophoretic mobility shift assay (EMSA) was performed using increasing concentrations of each scFv (Figure 1B). Both the WT (lanes 5–7) and D31N (lanes 12–14) scFv proteins were able to bind DNA, whereas the truncated scFv protein (lanes 2–4, 9–11, 16–18 and 23–25), the R92N protein (lanes 19–21), and the D31N/R92N protein (lanes 26–28) were unable to bind DNA. The D31N protein demonstrated higher DNA binding affinity than WT, consistent with previous work based on ELISA analysis (24,28).
Previously, Zack et al. correlated the ability of 3E10 to bind DNA with its cell-penetrating activity (28). To validate this finding and to compare intracellular localization of our scFv proteins, we treated human U2OS cells with each purified scFv protein by simple addition to the culture medium, followed by immunofluorescence microscopy using an antibody specific to the MBP tag. Both the WT and D31N scFv purified proteins were able to penetrate cells and localize primarily to the cytoplasm (Figure 1C). However, the truncated, R92N, and D31N/R92N purified proteins were unable to penetrate cells (Figure 1C). A summary of the DNA binding and cell penetration properties for each purified scFv is provided in Figure 1D.
3E10 inhibits HDR independently of DNA binding
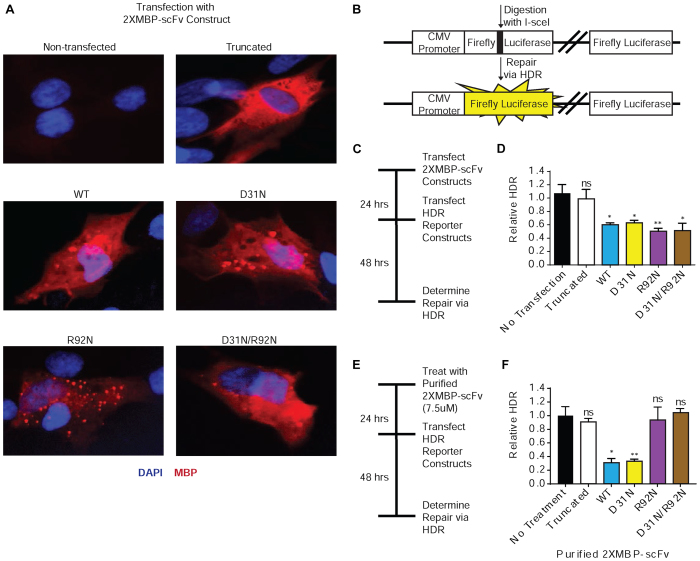
To assess the effect that each scFv variant has on HDR, a reporter gene assay was performed. Because three of the five scFvs are unable to penetrate cells as purified proteins added to cell culture medium, we expressed the recombinant scFv proteins intracellularly through transient transfection with the respective plasmid constructs. Figure 2A documents that all five scFv proteins are expressed upon transient transfection of the expression vectors into primary human skin fibroblasts, and further demonstrates primarily cytoplasmic localization. We then interrogated repair of an I-SceI nuclease-induced DSB within a luciferase reporter gene (29) in the presence of each expressed scFv. A schematic of the luciferase reporter construct used to assay HDR is provided in Figure 2B. Following generation of a DSB by I-SceI, restoration of functional luciferase requires HDR using the downstream luciferase gene fragment as a template. Cells were transfected with the scFv expression constructs and 24 h later with the I-SceI-digested luciferase reporter, as schematized in Figure 2C. Luciferase expression as a measure of HDR was determined 48 h later and normalized to transfection controls (Figure 2d and Supplemental Figure S1). As expected, the truncated scFv had no effect on DSB repair of the reporter construct by HDR. Surprisingly, all four of the non-truncated scFvs inhibited HDR to similar levels (∼40–50%). As the R92N scFvs cannot bind DNA, these results indicate that 3E10 inhibits HDR independently of its ability to bind DNA. Next, we tested the effect of each purified scFv variant on HDR in a protocol where cells were treated with the purified proteins by addition to the cell culture medium (as opposed to the experiment above in which the proteins were expressed intracellularly from transfected plasmids), followed by transfection with the I-SceI-digested luciferase reporter (Figure 2E). In this case, only the WT and D31N scFv were capable of inhibiting HDR; both reduced repair by >70% (Figure 2F). While the D31N/R92N and R92N scFvs inhibit HDR when expressed inside cells from a plasmid vector, the purified proteins fail to penetrate cells, and consequently, do not impact repair of the damaged plasmid (Figure 2F).
Figure 2.
3E10 scFv proteins that do not bind DNA still inhibit HDR. (A) Primary fibroblasts were transfected with the 2XMBP-scFv expression constructs. Cellular expression of each scFv was verified via immunofluorescence. The effect of each scFv on double strand break repair pathways was assayed using a luciferase plasmid-based host cell reactivation assay. (B) Schematic of the pre-digested luciferase constructs used to assay homology-directed repair (HDR). (C) Primary skin fibroblasts were transfected with 2XMBP-scFv expression constructs and then transfected with the pre-digested luciferase reporter constructs. (D) Results from the luciferase plasmid-based host cell reactivation assay, plotted as relative HDR (normalized to cells transfected with the luciferase constructs alone) after transfection with scFv expression constructs. (E) Primary skin fibroblasts were pre-treated with purified scFv proteins and then transfected with the pre-digested luciferase reporter constructs. (F) Results from the luciferase plasmid-based host cell reactivation assay, plotted as relative HDR (normalized to cells transfected with the luciferase constructs alone) after treatment with purified 2XMBP-scFv. Error bars represent the SEM; **P <0.01 and *P <0.05 by unpaired t-test.
To determine whether inhibition of DNA repair by 3E10 is specific to HDR, we tested the effect of scFv proteins (as well as the original full length 3E10 antibody) on NHEJ using a reporter assay similar to the one for HDR above (Supplemental Figure S2). Neither full-length 3E10 nor any of the scFvs affected NHEJ.
The finding that both of the R92N-containing scFv proteins fail to bind DNA and yet still inhibit HDR upon intracellular expression suggested that 3E10 acts through another cellular target other than DNA to impact HDR. To probe for this, we performed affinity pull-downs from lysates of HeLa cells expressing either the WT full length 3E10, the D31N full length 3E10 or empty vectors using protein A/G beads (to which the untagged full length 3E10 binds), followed by western blots for a panel of DNA repair factors (Supplemental Figure S3). Out of the 17 factors screened, the results identified RAD51 and RAD52 as putative targets. However, while protein pull-downs with purified RAD51 confirmed an interaction between RAD51 and 3E10 (see below) they failed to show any interaction between RAD52 and 3E10 (Supplemental Figure S3D). To confirm these results, we expressed the D31N scFv as a 2XMBP fusion protein in human 293T cells. Following binding to amylose beads, we washed the bound 2XMBP-scFv protein extensively, and then incubated the bead-protein complex with purified RAD51. As shown in Figure 3A (lane 5), RAD51 bound to the 3E10 D31N scFv but not to the 2XMBP tag alone (Figure 3A, lane 3) or to the amylose beads (Figure 3A, lane 2). The BRC1–8 region of BRCA2 was tested in parallel as a positive control for RAD51 binding and showed the expected physical interaction (Figure 3A, lane 7).
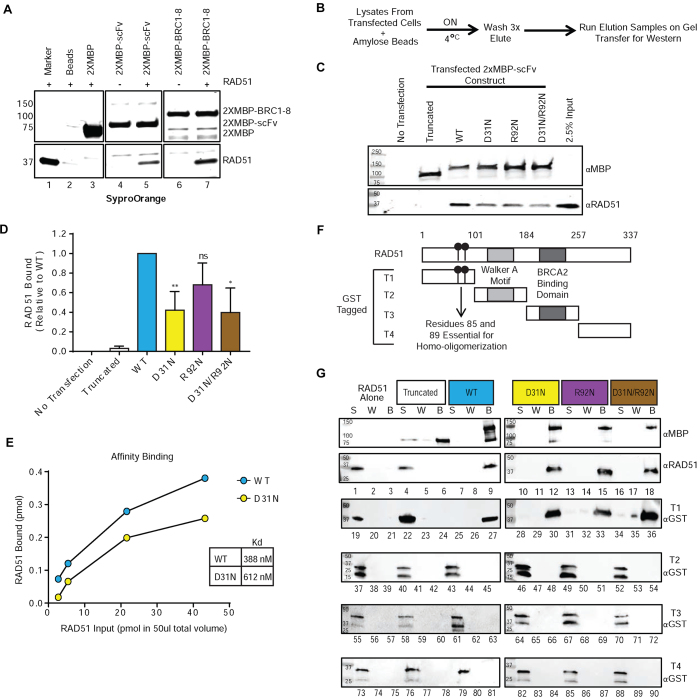
Figure 3.
3E10 physically interacts with the N-terminal domain of human RAD51. (A) Human 293T cells were transfected with expression constructs for the indicated 2XMBP fusion proteins, and cell lysates were batch bound to amylose beads, washed, and incubated in the presence or absence of purified RAD51. The bead complexes were then washed, eluted with maltose, and analyzed by SDS-PAGE. The gel was stained with SyproOrange to visualize proteins. Lane 1 is 1 μg of purified RAD51 as a marker. Lane 2 is purified RAD51 incubated with amylose beads alone. Lane 3 is 2XMBP tag alone bound to amylose beads and incubated with RAD51. (B) Protocol for an affinity pull-down performed using lysates of 293T cells left untransfected or transfected with the 2XMBP-scFv expression constructs. Western blot results from the affinity pull-down are shown in (C). (D) Quantification of the endogenous RAD51 bound to expressed 2XMBP-scFvs (combined levels from four experiments; normalized to MBP levels and then normalized to WT 2XMBP-scFv). (E) Affinity binding assay results depicting the amount of RAD51 pulled down with each 2XMBP-scFv plotted against the total amount of RAD51 entered into each pull-down reaction (2.7, 5.41, 21.62 and 43.24 pmol in a total volume of 50 μl). The binding curves were fit to a hyperbolic interpolation and the Kd values (nM) for the purified WT or D31N scFvs and purified RAD51 were calculated. (F). Diagram of the four purified fragments of human RAD51. (G) Protein pull downs were performed with purified scFv proteins and full-length purified human RAD51 and, the four purified fragments of human RAD51. The supernatant (S), wash (W) and bead elution (B) samples were interrogated for the presence of RAD51 by western blotting blot. Error bars represent the SEM; **P < 0.01, and *P < 0.05 by unpaired t-test.
As a next step, we performed affinity pull-downs from human 293T cells either untransfected or transfected with each of the five 2XMBP-scFv expression constructs (Figure 3B). This was performed to interrogate the intracellular interaction between 3E10 and RAD51 and to confirm that all four of the non-truncated 2XMBP-scFvs pulled down endogenous RAD51 from the 293T lysate. Western blots for RAD51 indicate that all four of the non-truncated variants of the 2XMBP-scFv pulled down endogenous RAD51 (Figure 3C). We noted that the expressed WT 2XMBP-scFv had the strongest interaction with RAD51, and that the presence of the D31N mutation slightly diminished the interaction with cellular RAD51. The levels of RAD51 bound (combined levels from four experiments; normalized to MBP levels and then normalized to WT 2XMBP-scFv) were plotted to depict this trend (Figure 3D).
Because the D31N mutation appeared to alter the interaction of the 3E10 scFv with cellular RAD51, we further evaluated the relative binding affinities of purified WT and D31N scFv proteins for purified RAD51 in vitro (29). Purified WT and D31N scFv proteins at a fixed concentration (400 ng in a final volume of 50 μl; 72.7 nM) were incubated with increasing amounts of purified RAD51 (100, 200, 800, 1600 ng; 54, 108, 432 and 864 nM, respectively), and the interacting complex was pulled down via the 2XMBP affinity tag on the scFvs using amylose resin. The bound complex was then washed and the protein was eluted from the amylose beads and run on a gel to evaluate the amount of RAD51 bound. The amount of RAD51 pulled down with 2XMBP-scFv proteins was determined using standard curves generated from known concentrations of RAD51 and 2XMBP-scFv proteins run in parallel (Supplemental Figures S4B and S4C), and the amount bound to the 2XMBP-scFv proteins was plotted against the amount of RAD51 titrated into the reaction to estimate the Kd values (Figure 3E and Supplemental Figure S4D). Consistent with the affinity pull-downs from human 293T cells, the WT 2XMBP-scFv had a stronger interaction (Kd = 388 nM) with RAD51 compared to the scFv with the D31N mutation (Kd = 612 nM).
3E10 physically interacts with the N-terminal domain of RAD51
To identify the domain of RAD51 to which the scFvs bind, we purified four fragments of RAD51 expressed in Escherichia coli cells using a GST tag (30). Fragment 1 (T1) is the N-terminal domain of RAD51, containing residues 86 and 89, which are essential for RAD51 homo-oligomerization and filament formation (31). Fragment T2 contains the Walker motifs essential for ATP-binding and nucleotide-binding (32). Fragment T3 contains the BRCA2-binding domain (30) and fragment T4 is the C-terminus of RAD51. These fragments are schematized in Figure 3f.
Affinity pull-downs were then performed with full length RAD51 as a control, and each of the four purified fragments of RAD51, representing separate domains of the protein. The scFvs were immobilized on amylose resin, incubated with purified RAD51 (either full length or fragment), washed, and then eluted. The supernatant (S), wash (W) and bead elution (B) samples were interrogated for the presence of RAD51 by western blotting (Figure 3G). The results show that full length RAD51 was pulled down by all four non-truncated scFv proteins (Figure 3G, lanes 9, 12, 15 and 18). As controls, when RAD51 was incubated with amylose beads alone or with the truncated 2XMBP-scFv, RAD51 was found only in the supernatant (Figure 3G, lanes 1 and 4), indicating lack of an interaction with the beads alone or the truncated scFv protein. Taken together, these results indicate that 3E10 specifically interacts with RAD51. As shown in Figure 3G, the T1 fragment of RAD51 was also present in the eluate (B) for the four non-truncated purified scFvs, indicating a physical interaction with the respective scFv occurred (Figure 3G, lanes 27, 30, 33 and 36). When T1 of RAD51 was incubated with amylose beads alone or the truncated 2xMBP-scFv, T1 was located only in the supernatant (S), indicating there was no physical interaction (Figure 3G, lanes 19 and 22). There was no physical interaction between any of the other fragments of RAD51 and any of the scFvs (Figure 3G, lanes 37–90), revealing that the 3E10 scFv specifically interacts with the N-terminal T1 domain of RAD51.
3E10 inhibits RAD51 accumulation on DNA
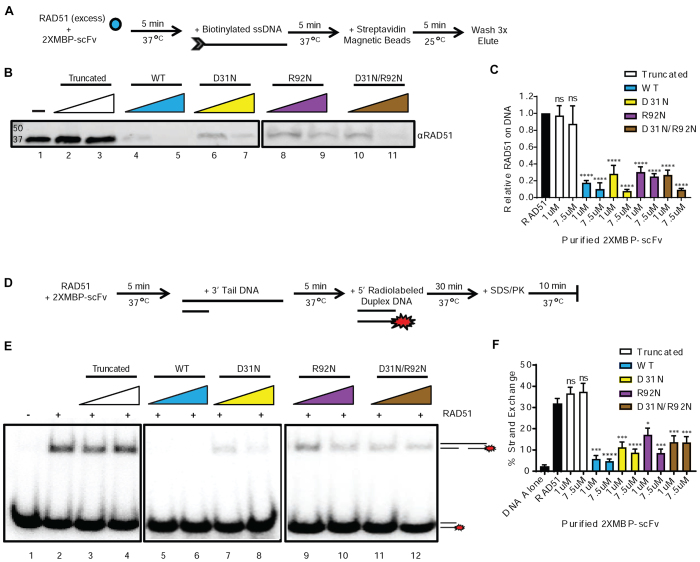
Because the N-terminal domain of RAD51 contains residues that are essential for RAD51 homo-oligomerization and filament formation (31), we hypothesized that an interaction between the 3E10 scFv and the N-terminal domain of RAD51 would inhibit RAD51 binding to DNA. RAD51 and the 3E10 scFvs were pre-incubated, followed by addition of the biotinylated ssDNA, as schematized in Figure 4A. We found that all four non-truncated scFvs showed direct inhibition of RAD51 accumulation on ssDNA (Figure 4B and C). Although the DNA binding competition between RAD51 and either the WT or the D31N scFv may partially contribute the activity of these two variants, the observation that all four non-truncated scFvs significantly reduced RAD51’s ability to bind to the ssDNA suggest that 3E10 directly affects RAD51 loading on ssDNA through a physical interaction with RAD51, and not primarily through DNA binding that could in theory compete with occupancy of RAD51 on ssDNA tails (since the R92N variants do not bind DNA).
Figure 4.
3E10 inhibits RAD51 binding to DNA and RAD51-mediated strand exchange. (A) Scheme of RAD51 DNA binding assay performed with pre-incubation of RAD51 and 2XMBP-scFv proteins. (B) Representative assay results. (C) Quantification of RAD51 bound to DNA for each condition. (D) A strand exchange assay was performed with pre-incubation of RAD51 and 2XMBP-scFv proteins. (E) Representative assay results. (F) Quantification of RAD51 mediated strand exchange for each condition. Error bars represent the SEM; ****P < 0.0001, ***P < 0.001, **P < 0.01 and *P < 0.05 by unpaired t-test.
3E10 inhibits RAD51-mediated strand exchange
3E10 was previously shown to inhibit RAD51 mediated strand exchange (26), but this was attributed to its DNA binding affinity. Based on the new evidence presented above showing that 3E10 physically interacts with RAD51, and that the 3E10 R92N scFvs that cannot bind DNA still interact with RAD51, we next tested the ability of each of the 3E10 scFvs to inhibit RAD51 mediated strand exchange.
RAD51 and the scFv proteins were pre-incubated before addition of the DNA molecules, as schematized in Figure 4d. Because all four non-truncated scFvs substantially inhibited nucleation of RAD51 on ssDNA in the filament formation assay (Figure 4B and C), we hypothesized that pre-incubation of the non-truncated scFv proteins with RAD51 would also inhibit strand exchange in this experimental setup. Representative assay results are shown in Figure 4e and the quantification of RAD51 mediated strand exchange for each condition is shown in Figure 4F. All four non-truncated scFv proteins greatly reduced RAD51-mediated strand exchange, supporting the finding that the 3E10 scFv reduces RAD51 loading on ssDNA through a physical interaction with RAD51, which functionally reduces RAD51’s ability to facilitate homology dependent strand exchange.
3E10 inhibits RAD51 nuclear localization and foci formation
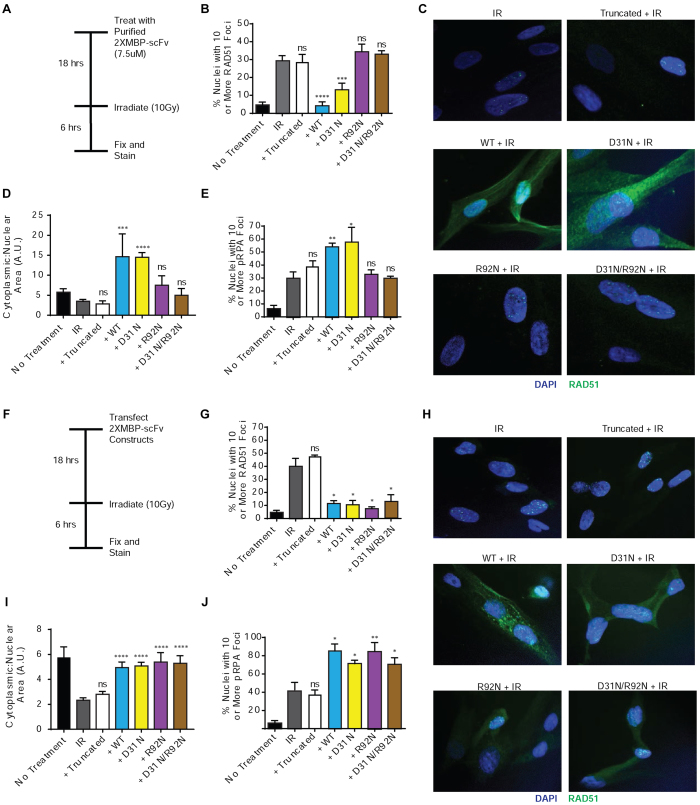
We next tested the impact of the 3E10 scFvs on the formation of nuclear RAD51 foci cells exposed to DNA damaging agents as a measure of RAD51’s participation in DNA repair. Primary skin fibroblasts were left untreated or treated with purified scFv proteins, then irradiated with 10 Gy IR, and subsequently fixed for immunofluorescence microscopy using an anti-RAD51 antibody (Figure 5A). Cells with 10 or more RAD51 foci were scored for each condition (Figure 5B). WT and D31N scFvs both strongly reduced RAD51 foci formation. The WT version had a slightly larger effect. The other scFvs, which cannot penetrate cells, had no effect. Representative images of treated primary skin fibroblasts are shown in Figure 5C and Supplemental Figure S5. Interestingly, in these images we noted that the WT and D31N scFvs not only inhibited RAD51 foci formation in the nucleus, but also caused substantial retention of RAD51 in the cytoplasm. The extent of cytoplasmic retention was quantified using image analysis software (Figure 5D).
Figure 5.
3E10 inhibits RAD51 nuclear localization and foci formation after irradiation. (A) Primary skin fibroblasts were left untreated or pretreated with purified 2XMBP-scFv proteins and then irradiated with 10Gy IR and fixed for immunofluorescence. (B) Cells with 10 or more RAD51 foci were scored for each condition. (C) Representative images of treated primary skin fibroblasts. The cytoplasmic fraction of RAD51 was also scored for each condition (D), as well as cells with 10 or more RPA foci (E). (F) Primary skin fibroblasts were also transfected with 2XMBP-scFv expression constructs and then irradiated with 10Gy IR and fixed for immunofluorescence. (G) Cells with 10 or more RAD51 foci were scored for each condition. (H) Representative images of transfected primary skin fibroblasts. The cytoplasmic fraction of RAD51 (I) was also scored, in addition to cells with 10 or more RPA foci (J). Error bars represent the SEM; ****P < 0.0001, ***P < 0.001, **P < 0.01, and *P < 0.05 by unpaired t-test.
Under normal conditions, RAD51 can be observed in both the cytoplasm and nucleus of cells. It is thought that under cellular stress and DNA damage, RAD51 is transported to the nucleus and loaded onto DNA by BRCA2 (29). RAD51C, RAD52 and BRCA2 have all been suggested to play a role in the translocation of RAD51 from the cytoplasm to the nucleus although the exact mechanism is yet to be determined (29,33,34). Therefore, we hypothesize that, since the 2XMBP-scFvs are primarily localized in the cytoplasm, the interaction between RAD51 and the 3E10 scFvs likely occurs in the cytoplasm. This may interfere with RAD51’s interaction with other cellular proteins which may in turn affect RAD51 transport to the nucleus upon DNA damage.
After a DNA DSB is resected to create a ssDNA overhang, the ssDNA tail is coated with Replication Protein A (RPA) to prevent additional nuclease digestion (8). This RPA filament is displaced by RAD51. To test the hypothesis that 3E10 scFv's inhibition of RAD51 foci formation would lead to an accumulation or at least persistence of RPA at sites of DNA DSBs, transfected cells were fixed and stained for phospho-RPA (the activated form) after exposure to 10 Gy IR. We found that treatment of cells with either the WT or D31N scFv proteins led to a slight but statistically significant increase in pRPA foci as compared to irradiation alone (Figure 5e and Supplemental Figure S6). The other three scFv proteins, which do not penetrate cells in purified form, did not affect RPA foci.
To test the impact of the scFvs that cannot penetrate cells, primary fibroblasts were also transfected with the 2XMBP-scFv expression constructs and then irradiated with 10 Gy IR and fixed for immunofluorescence at two different time points (Figure 5f and Supplemental Figure S7A). Again, cells with 10 or more RAD51 foci were scored for each condition (Figure 5G and Supplemental Figure S7B). As expected, the truncated scFv had no effect on RAD51 localization or foci formation. However, at both time points, all four of the other transfected scFvs significantly impaired RAD51 nuclear localization and foci formation after exposure to IR. Representative images of transfected primary skin fibroblasts are shown in Figure 5G and Supplemental Figure S8. As above, the cytoplasmic retention of RAD51 was also quantified using image analysis software (Figure 5H and Supplemental Figure S7C), and cells with 10 or more RPA foci were scored at two time-points post transfection (Figure 5I and Supplemental Figure S7D and Supplemental Figure S9). Again, transfection with the truncated scFv had no effect RAD51 localization or on RPA foci formation. However, at both time points, all four of the other transfected scFvs led to increased cytoplasmic retention of RAD51 and to a significant increase in RPA foci after exposure to IR. For comparison, we also found that treatment of cells with the full-length 3E10 similarly impaired RAD51 foci formation after exposure to IR (Supplemental Figure S10).
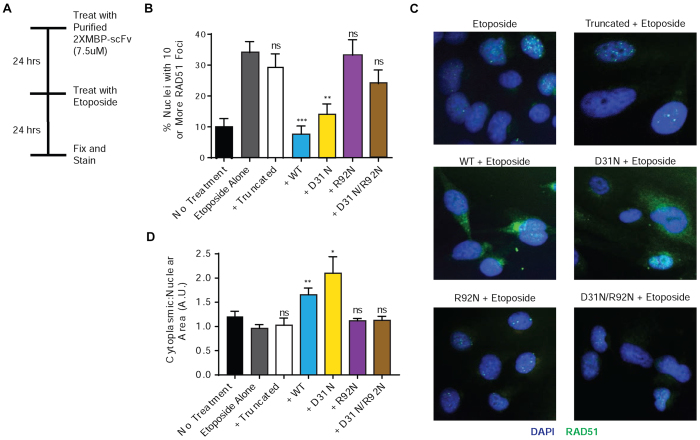
Next, we probed the impact of the 3E10 scFvs on RAD51 nuclear localization and foci formation in response to a topoisomerase inhibitor, etoposide, another well-established inducer of RAD51 foci (Figure 6a). In this experiment, cells were treated with the purified 3E10 scFv proteins, and then exposed to etoposide for 24 h. Cells were fixed and stained, and the number of cells with 10 or more RAD51 foci and the proportion of cytoplasmic RAD51 were scored for each condition (Figure 6B and D, respectively, with representative images in Figure 6C and Supplemental Figure S11). Treatment with the WT and D31N scFv proteins both led to a decrease in RAD51 nuclear localization and foci formation. The other purified scFvs had no effect, again in keeping with lack of cell penetration.
Figure 6.
3E10 inhibits RAD51 nuclear localization and foci formation after treatment with etoposide. (A) U2OS cells were left untreated or pretreated with purified 2xMBP-3E10 scFv proteins and then treated with etoposide and fixed for immunofluorescence. (B) Cells with 10 or more RAD51 foci were scored for each condition. (C) Representative images of treated U2OS cells. (D) The cytoplasmic fraction of RAD51 was also scored for each condition. Error bars represent the SEM; ***P < 0.001, **P < 0.01 and *P < 0.05 by unpaired t-test.
The strength of the 3E10-RAD51 interaction affects the extent of chemosensitization in BRCA2-deficient and PTEN-deficient cells
Because of the differential affinity of the various 2xMBP-scFvs to RAD51 and DNA, we evaluated the effect of the scFvs on cell survival in the context if DNA damage. BRCA2 and PTEN are both known to play roles in DSB repair, and cells deficient in either of these proteins have been reported to have elevated levels of unrepaired DNA damage (4,5,29,35,36). Previous publications have reported that 3E10 is synthetically lethal with BRCA2-deficiency as well as PTEN-deficiency (26,37). Based on this, we elected to assess chemosensitization to Etoposide in BRCA2-deficient PEO-1 cells and PTEN-deficient U251 cells. To evaluate whether DNA binding mutations of the 2XMBP-scFvs alter the sensitivity of these cells to Etoposide independent of cell penetration ability, PEO-1 and U251 cells were transfected with the 2XMBP-scFv expression constructs and then reseeded into Etoposide for clonogenic survival assays. Western blot results confirming the expression of the 2XMBP-scFvs are shown in Supplemental Figure S12a and c. In both the transfected PEO-1 and U251 cells, expression of the WT and R92N 2XMBP-scFvs yielded enhanced cytotoxicity to Etoposide compared to the D31N 2XMBP-scFv (Supplemental Figure S12B and D). These data provide evidence that the 3E10-RAD51 interaction is more important for chemosensitization in cells then the 3E10–DNA interaction.
Next, we assessed the differential effect of treatment of cells with the purified WT and D31N scFvs (as opposed to transfection with the expression constructs). U251 cells were pretreated with the 2XMBP-scFv purified proteins and then reseeded into Etoposide for clonogenic survival assays. Here, both the WT and D31N scFvs sensitized the U251 cells to Etoposide to comparable levels, showing that the increased DNA binding and thereby cellular uptake conferred by D31N compensates for its reduced RAD51 interaction in terms of overall cellular activity for sensitization to Etoposide (Supplemental Figure S12E).
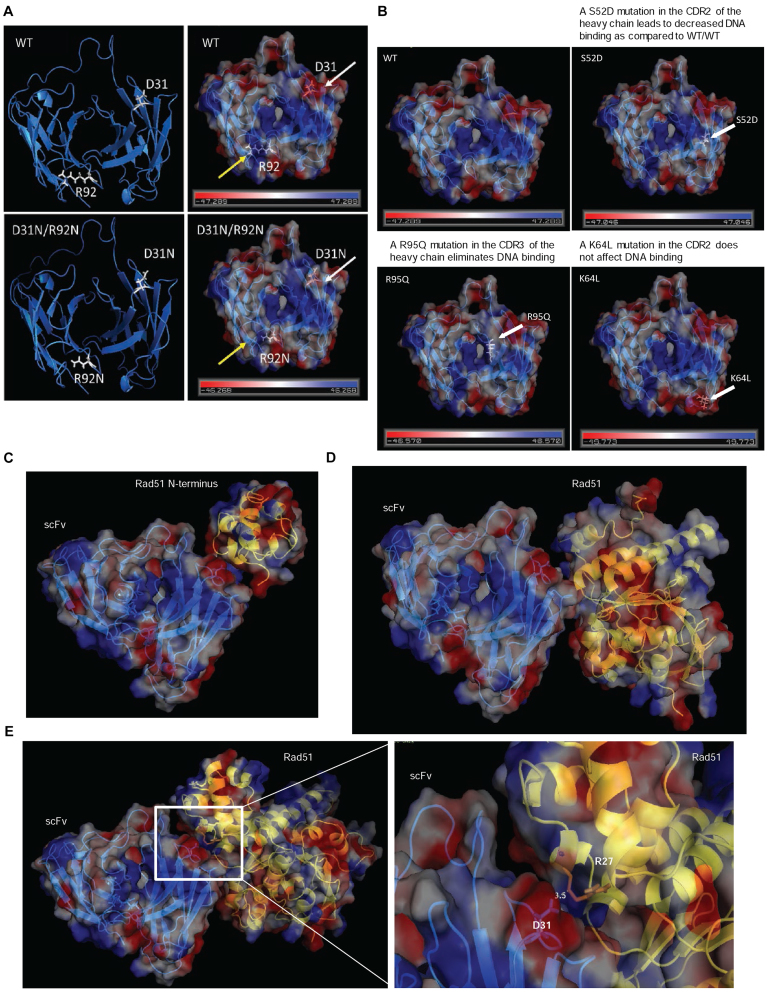
Structural modeling suggests a DNA binding pocket and RAD51 docking site
A predicted structure of WT 3E10 scFv was generated using SWISS MODEL (Figure 7A) (38,39). An electrostatic map was then generated using PyMOL to assess structural regions which might confer a DNA binding pocket (Figure 7A) (PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC.). This electrostatic map indicates that the D31N mutation results in an enhanced positively charged region in the predicted DNA binding pocket (yellow arrows), while the R92N mutation eliminates a positively charged region in the predicted DNA binding pocket (white arrows). This analysis supports the finding that the D31N scFv has enhanced DNA binding over the WT scFv and may explain why this DNA binding ability is lost when the R92N point mutation is introduced. In order to further validate the predicted overall structure and DNA binding pocket, additional point mutations that have been previously published as having an effect on 3E10 scFv's DNA binding ability (24,28) were mapped on the generated predicted structure of the 3E10 scFv (Figure 7B). We found that these other point mutations that were shown to disrupt DNA binding also altered the electrostatic profile of the predicted DNA binding pocket in manner expected to reduce affinity for DNA.
Figure 7.
A molecular model of the 3E10 scFv supports experimental evidence of the DNA binding pocket and the interaction with human RAD51. (A) An electrostatic map generated from a predicted structure of the 3E10 scFv. The D31N mutation is marked with white arrows, while the R92N mutation is marked by yellow arrows. (B) Additional point mutations that have been previously published as having an effect on 3E10 scFv's DNA binding ability were plotted on the generated predicted structure of the 3E10 scFv. (C) A potential model of the interaction between the N-terminus of RAD51 and the 3E10 scFv. (D) A potential model of the interaction between the human RAD51-BRCA2 BRC repeat complex (PDB 1N0W) and the 3E10 scFv. (E) A potential model of the interaction between the Saccharomyces cerevisiae RAD51 structure (PDB 3LDA) and the 3E10 scFv. RAD51 is represented by the yellow ribbon structure in C–E.
Next, we modeled the putative WT 3E10 scFv–RAD51 interaction using the Vakser Lab Protein–Protein Docking Web Server from the Center for Computational Biology at the The University of Kansas (40,41). Further analysis and simulation was performed using PyMOL. A preliminary interaction model between the N-terminal domain of RAD51 and WT 3E10 scFv is provided in Figure 7C (PDB 1B22 (42)). Additional models were generated using the human RAD51-BRCA2 BRC repeat complex (PDB 1N0W (31) shown in Figure 7D) as well as the Saccharomyces cerevisiae RAD51 structure (PDB 3LDA (43) shown in Figure 7E). The model shown in Figure 7E places the D31 residue in the heavy chain at the interface of the 3E10 scFv-RAD51 interaction. This would be consistent with our observation that the D31N mutation partially reduces the extent of the 3E10/RAD51 interaction (Figure 3D and E).
DISCUSSION
The results presented here demonstrate that 3E10 inhibits HDR in a manner that is independent of its ability to bind DNA and instead depends on its direct interaction with RAD51. Through a cell-based assay for HDR, we found that variants of the scFv of 3E10 (R92N and D31N/R92N) that cannot bind DNA were still able to inhibit HDR at a DNA double-strand break. This raised the possibility that 3E10 inhibits HDR independently of its DNA binding ability. In a candidate survey for interactions of 3E10 with DNA repair proteins by pull downs from cell lysates, we found initial evidence that that 3E10 physically interacts with endogenous RAD51. Further analysis with purified RAD51 fragments revealed that 3E10 specifically binds to the N-terminal domain of RAD51. This domain contains residues previously reported to be essential for RAD51 homo-oligomerization that promotes its nucleation on single-stranded DNA tails (31). The resulting formation of a RAD51 nucleofilament is a critical step in the resolution of a DNA DSB through HDR. In assays with purified RAD51 and 3E10 scFvs, we found that, regardless of DNA binding ability, each of the 3E10 scFvs inhibit the accumulation of RAD51 on ssDNA. Consistent with these results, we found the scFvs hinder RAD51-mediated strand exchange, again independently of their ability to bind DNA. In cells treated with either IR or etoposide, DNA damaging agents known to provoke RAD51 mediated repair, treatment of cells with the WT and D31N 3E10 scFvs yielded substantial reduction in RAD51 foci formation and a notable increase in cytoplasmic localization of RAD51. Although the R92N scFvs had no effect when used to treat cells, when they were expressed intracellularly from expression constructs, they similarly impaired foci formation and increased cytoplasmic retention.
In regard to cancer therapy, it has been proposed that oncogene-induced replication stress, coupled with abnormalities in DNA damage checkpoints, renders cancers vulnerable to strategies that target DNA repair (44). In addition, many malignancies show over expression of RAD51 (16–18) and develop ‘addiction’ to high RAD51 levels, further pointing to RAD51 as a promising therapeutic target (15). Plus, numerous malignancies have relative deficiencies in HDR due to mutations in or reduced expression of BRCA1, BRCA2 and other HDR pathway factors (45,46), potentially rendering them susceptible to further inhibition of HDR capacity. Tumor hypoxia also impacts DNA repair capacity (47), and recent clinical trials have suggested that induced hypoxia (via treatment with cediranib, an angiogenesis inhibitor), can sensitize tumors to DNA repair inhibition (48,49). In light of this, our results showing that 3E10 directly inhibits RAD51 activity may provide a mechanistic basis to anticipate a therapeutic gain in the use of 3E10 in cancer therapy. Interestingly, while 3E10 is capable of penetrating most cells, tissues with high cell turnover and excessive extracellular DNA, such as tumors, show increased uptake of 3E10 (50). This property may further contribute to an increased therapeutic window, further highlighting the clinical potential of this unusual cell-penetrating antibody. Recent work by Noble et al suggests that engineering recombinant 3E10 molecules to comprise multiply valent scFvs can substantially boost activity (37). In this regard, the work presented here not only identifies 3E10 as a novel RAD51 inhibitor but also may help to guide further modifications to the 3E10 scFv to tune its DNA and RAD51 binding properties as a means to further increase efficacy for cancer therapy.
DATA AVAILABILITY
The following software programs were used:
GraphPad Prism (https://www.graphpad.com/scientific-software/prism/#6)
ImageJ (https://imagej.nih.gov/ij/index.html)
SWISS-MODEL (https://swissmodel.expasy.org/)
PyMol (https://www.pymol.org/)
Vakser Lab Protein-Protein Docking Web Server (http://vakser.compbio.ku.edu/resources/gramm/grammx/)
Known structures for RAD51 that were used to create a preliminary interaction model between the N-terminal domain of RAD51 and WT 3E10 scFv. These RAD51 structures have been deposited with the Protein Data bank by other groups under the accession numbers PDB 1B22, PDB 1N0W, and PDB 3LDA.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank Olga Buzovetsky (Yale University) for her guidance in operating PyMOL and Weixing Zhao and Patrick Sung (Yale University) for their assistance in purifying fragments of RAD51. We also would like to thank Dr Alexander MacKerell at the University of Maryland for his helpful suggestions regarding protein docking prediction modeling.
Author contributions: A.T., R.B.J. and P.M.G. designed the experiments. A.T., D.C.H. and R.B.J. performed the experiments. A.T., D.C.H., R.B.J. and P.M.G. analyzed and interpreted the data. A.T., R.B.J. and P.M.G. contributed to the preparation of the figures and wrote the manuscript.
Footnotes
Present address: Peter M. Glazer, Yale University School of Medicine, New Haven, CT 06510, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [R35CA197574, R01CA168733, R01ES005775 to P.M.G.]; National Institutes of Health Training Grants [GM007324, T32 HD007149 to Yale University for A.T.]; American Cancer Society [#IRG 58-012-55 to R.B.J.]; Breast Cancer Alliance [to R.B.J.]; Pilot Project Program grant from Women's Health Research at Yale [to R.B.J.]; Liz Tilberis Early Career Award from the Ovarian Cancer Research Fund Alliance [to R.B.J.]; Yale Cancer Center pilot grant [to R.B.J.]. Funding for open access charge: NIH.
Conflict of interest statement. P.M.G. is an inventor on a patent application filed by Yale University covering the use of the 3E10 antibody for cancer therapy.
REFERENCES
- 1. Scott A.M., Wolchok J.D., Old L.J.. Antibody therapy of cancer. Nat. Rev. Cancer. 2012; 12:278–287. [DOI] [PubMed] [Google Scholar]
- 2. Pillay V., Allaf L., Wilding A.L., Donoghue J.F., Court N.W., Greenall S.A., Scott A.M., Johns T.G.. The plasticity of oncogene addiction: implications for targeted therapies directed to receptor tyrosine kinases. Neoplasia. 2009; 11:448–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Scott A.M., Allison J.P., Wolchok J.D.. Monoclonal antibodies in cancer therapy. Cancer Immun. 2012; 12:14. [PMC free article] [PubMed] [Google Scholar]
- 4. Bryant H.E., Schultz N., Thomas H.D., Parker K.M., Flower D., Lopez E., Kyle S., Meuth M., Curtin N.J., Helleday T.. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005; 434:913–917. [DOI] [PubMed] [Google Scholar]
- 5. Farmer H., McCabe N., Lord C.J., Tutt A.N., Johnson D.A., Richardson T.B., Santarosa M., Dillon K.J., Hickson I., Knights C. et al. . Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005; 434:917–921. [DOI] [PubMed] [Google Scholar]
- 6. Klein T.J., Glazer P.M.. The tumor microenvironment and DNA repair. Semin. Radiat. Oncol. 2010; 20:282–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Du L., Zhou L.J., Pan X.J., Wang Y.X., Xu Q.Z., Yang Z.H., Wang Y., Liu X.D., Zhu M.X., Zhou P.K.. Radiosensitization and growth inhibition of cancer cells mediated by an scFv antibody gene against DNA-PKcs in vitro and in vivo. Radiat. Oncol. 2010; 5:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. San Filippo J., Sung P., Klein H.. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008; 77:229–257. [DOI] [PubMed] [Google Scholar]
- 9. Huertas P. DNA resection in eukaryotes: deciding how to fix the break. Nat. Struct. Mol. Biol. 2010; 17:11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sung P., Klein H.. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat. Rev. Mol. Cell Biol. 2006; 7:739–750. [DOI] [PubMed] [Google Scholar]
- 11. Beernink H.T., Morrical S.W.. RMPs: recombination/replication mediator proteins. Trends Biochem. Sci. 1999; 24:385–389. [DOI] [PubMed] [Google Scholar]
- 12. Yang H., Li Q., Fan J., Holloman W.K., Pavletich N.P.. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature. 2005; 433:653–657. [DOI] [PubMed] [Google Scholar]
- 13. Jensen R.B., Carreira A., Kowalczykowski S.C.. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010; 467:678–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chi P., San Filippo J., Sehorn M.G., Petukhova G.V., Sung P.. Bipartite stimulatory action of the Hop2-Mnd1 complex on the Rad51 recombinase. Genes Dev. 2007; 21:1747–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mason J.M., Logan H.L., Budke B., Wu M., Pawlowski M., Weichselbaum R.R., Kozikowski A.P., Bishop D.K., Connell P.P.. The RAD51-stimulatory compound RS-1 can exploit the RAD51 overexpression that exists in cancer cells and tumors. Cancer Res. 2014; 74:3546–3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Klein H.L. The consequences of Rad51 overexpression for normal and tumor cells. DNA Repair (Amst.). 2008; 7:686–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Maacke H., Opitz S., Jost K., Hamdorf W., Henning W., Kruger S., Feller A.C., Lopens A., Diedrich K., Schwinger E. et al. . Over-expression of wild-type Rad51 correlates with histological grading of invasive ductal breast cancer. Int. J. Cancer. 2000; 88:907–913. [DOI] [PubMed] [Google Scholar]
- 18. Mitra A., Jameson C., Barbachano Y., Sanchez L., Kote-Jarai Z., Peock S., Sodha N., Bancroft E., Fletcher A., Cooper C. et al. . Overexpression of RAD51 occurs in aggressive prostatic cancer. Histopathology. 2009; 55:696–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Takenaka T., Yoshino I., Kouso H., Ohba T., Yohena T., Osoegawa A., Shoji F., Maehara Y.. Combined evaluation of Rad51 and ERCC1 expressions for sensitivity to platinum agents in non-small cell lung cancer. Int. J. Cancer. 2007; 121:895–900. [DOI] [PubMed] [Google Scholar]
- 20. Connell P.P., Jayathilaka K., Haraf D.J., Weichselbaum R.R., Vokes E.E., Lingen M.W.. Pilot study examining tumor expression of RAD51 and clinical outcomes in human head cancers. Int. J. Oncol. 2006; 28:1113–1119. [PubMed] [Google Scholar]
- 21. Marsden C.G., Jensen R.B., Zagelbaum J., Rothenberg E., Morrical S.W., Wallace S.S., Sweasy J.B.. The tumor-associated variant RAD51 G151D induces a hyper-recombination phenotype. PLoS Genet. 2016; 12:e1006208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Alarcon-Segovia D. Antinuclear antibodies: to penetrate or not to penetrate, that was the question. Lupus. 2001; 10:315–318. [DOI] [PubMed] [Google Scholar]
- 23. Weisbart R.H., Stempniak M., Harris S., Zack D.J., Ferreri K.. An autoantibody is modified for use as a delivery system to target the cell nucleus: therapeutic implications. J. Autoimmun. 1998; 11:539–546. [DOI] [PubMed] [Google Scholar]
- 24. Zack D.J., Stempniak M., Wong A.L., Taylor C., Weisbart R.H.. Mechanisms of cellular penetration and nuclear localization of an anti-double strand DNA autoantibody. J. Immunol. 1996; 157:2082–2088. [PubMed] [Google Scholar]
- 25. Hansen J.E., Tse C.M., Chan G., Heinze E.R., Nishimura R.N., Weisbart R.H.. Intranuclear protein transduction through a nucleoside salvage pathway. J. Biol. Chem. 2007; 282:20790–20793. [DOI] [PubMed] [Google Scholar]
- 26. Hansen J.E., Chan G., Liu Y., Hegan D.C., Dalal S., Dray E., Kwon Y., Xu Y., Xu X., Peterson-Roth E. et al. . Targeting cancer with a lupus autoantibody. Sci. Transl. Med. 2012; 4:157ra142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jensen R. Purification of recombinant 2XMBP tagged human proteins from human cells. Methods Mol. Biol. 2014; 1176:209–217. [DOI] [PubMed] [Google Scholar]
- 28. Zack D.J., Yamamoto K., Wong A.L., Stempniak M., French C., Weisbart R.H.. DNA mimics a self-protein that may be a target for some anti-DNA antibodies in systemic lupus erythematosus. J. Immunol. 1995; 154:1987–1994. [PubMed] [Google Scholar]
- 29. Chatterjee G., Jimenez-Sainz J., Presti T., Nguyen T., Jensen R.B.. Distinct binding of BRCA2 BRC repeats to RAD51 generates differential DNA damage sensitivity. Nucleic Acids Res. 2016; 44:5256–5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Buisson R., Dion-Cote A.M., Coulombe Y., Launay H., Cai H., Stasiak A.Z., Stasiak A., Xia B., Masson J.Y.. Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination. Nat. Struct. Mol. Biol. 2010; 17:1247–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pellegrini L., Yu D.S., Lo T., Anand S., Lee M., Blundell T.L., Venkitaraman A.R.. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature. 2002; 420:287–293. [DOI] [PubMed] [Google Scholar]
- 32. Chen J., Morrical M.D., Donigan K.A., Weidhaas J.B., Sweasy J.B., Averill A.M., Tomczak J.A., Morrical S.W.. Tumor-associated mutations in a conserved structural motif alter physical and biochemical properties of human RAD51 recombinase. Nucleic Acids Res. 2015; 43:1098–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gildemeister O.S., Sage J.M., Knight K.L.. Cellular redistribution of Rad51 in response to DNA damage: novel role for Rad51C. J. Biol. Chem. 2009; 284:31945–31952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Feng Z., Scott S.P., Bussen W., Sharma G.G., Guo G., Pandita T.K., Powell S.N.. Rad52 inactivation is synthetically lethal with BRCA2 deficiency. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:686–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McEllin B., Camacho C.V., Mukherjee B., Hahm B., Tomimatsu N., Bachoo R.M., Burma S.. PTEN loss compromises homologous recombination repair in astrocytes: implications for glioblastoma therapy with temozolomide or poly(ADP-ribose) polymerase inhibitors. Cancer Res. 2010; 70:5457–5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bassi C., Ho J., Srikumar T., Dowling R.J., Gorrini C., Miller S.J., Mak T.W., Neel B.G., Raught B., Stambolic V.. Nuclear PTEN controls DNA repair and sensitivity to genotoxic stress. Science. 2013; 341:395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Noble P.W., Chan G., Young M.R., Weisbart R.H., Hansen J.E.. Optimizing a lupus autoantibody for targeted cancer therapy. Cancer Res. 2015; 75:2285–2291. [DOI] [PubMed] [Google Scholar]
- 38. Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Gallo Cassarino T., Bertoni M., Bordoli L. et al. . SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014; 42:W252–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Arnold K., Bordoli L., Kopp J., Schwede T.. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006; 22:195–201. [DOI] [PubMed] [Google Scholar]
- 40. Tovchigrechko A., Vakser I.A.. Development and testing of an automated approach to protein docking. Proteins. 2005; 60:296–301. [DOI] [PubMed] [Google Scholar]
- 41. Tovchigrechko A., Vakser I.A.. GRAMM-X public web server for protein-protein docking. Nucleic Acids Res. 2006; 34:W310–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Aihara H., Ito Y., Kurumizaka H., Yokoyama S., Shibata T.. The N-terminal domain of the human Rad51 protein binds DNA: structure and a DNA binding surface as revealed by NMR. J. Mol. Biol. 1999; 290:495–504. [DOI] [PubMed] [Google Scholar]
- 43. Chen J., Villanueva N., Rould M.A., Morrical S.W.. Insights into the mechanism of Rad51 recombinase from the structure and properties of a filament interface mutant. Nucleic Acids Res. 2010; 38:4889–4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Helleday T. Homologous recombination in cancer development, treatment and development of drug resistance. Carcinogenesis. 2010; 31:955–960. [DOI] [PubMed] [Google Scholar]
- 45. Powell S.N., Kachnic L.A.. Therapeutic exploitation of tumor cell defects in homologous recombination. Anticancer Agents Med. Chem. 2008; 8:448–460. [DOI] [PubMed] [Google Scholar]
- 46. Stoppa-Lyonnet D. The biological effects and clinical implications of BRCA mutations: where do we go from here?. Eur. J. Hum. Genet. 2016; 24(Suppl. 1):S3–S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Scanlon S.E., Glazer P.M.. Multifaceted control of DNA repair pathways by the hypoxic tumor microenvironment. DNA Repair (Amst.). 2015; 32:180–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lee J.M., Trepel J.B., Choyke P., Cao L., Sissung T., Houston N., Yu M., Figg W.D., Turkbey I.B., Steinberg S.M. et al. . CECs and IL-8 have prognostic and predictive utility in patients with recurrent platinum-sensitive ovarian cancer: biomarker correlates from the randomized phase-2 trial of olaparib and cediranib compared with olaparib in recurrent platinum-sensitive ovarian cancer. Front. Oncol. 2015; 5:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liu J.F., Tolaney S.M., Birrer M., Fleming G.F., Buss M.K., Dahlberg S.E., Lee H., Whalen C., Tyburski K., Winer E. et al. . A Phase 1 trial of the poly(ADP-ribose) polymerase inhibitor olaparib (AZD2281) in combination with the anti-angiogenic cediranib (AZD2171) in recurrent epithelial ovarian or triple-negative breast cancer. Eur. J. Cancer. 2013; 49:2972–2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Weisbart R.H., Chan G., Jordaan G., Noble P.W., Liu Y., Glazer P.M., Nishimura R.N., Hansen J.E.. DNA-dependent targeting of cell nuclei by a lupus autoantibody. Sci. Rep. 2015; 5:12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The following software programs were used:
GraphPad Prism (https://www.graphpad.com/scientific-software/prism/#6)
ImageJ (https://imagej.nih.gov/ij/index.html)
SWISS-MODEL (https://swissmodel.expasy.org/)
PyMol (https://www.pymol.org/)
Vakser Lab Protein-Protein Docking Web Server (http://vakser.compbio.ku.edu/resources/gramm/grammx/)
Known structures for RAD51 that were used to create a preliminary interaction model between the N-terminal domain of RAD51 and WT 3E10 scFv. These RAD51 structures have been deposited with the Protein Data bank by other groups under the accession numbers PDB 1B22, PDB 1N0W, and PDB 3LDA.