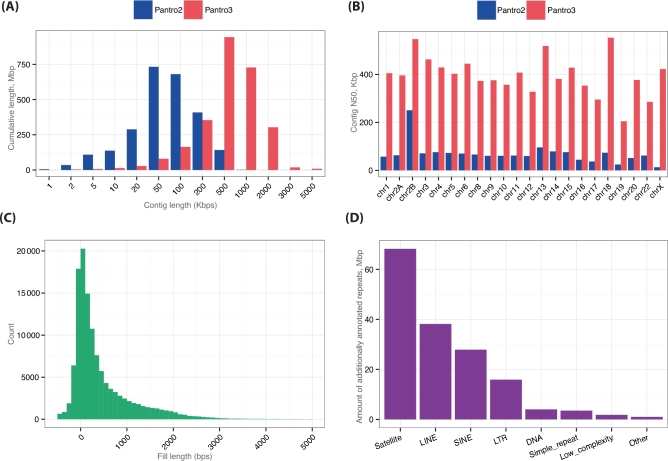
Figure 1:
(A) Genome-wide distribution of contig lengths between Pan_tro_2.1.4 and Pan_tro_3.0. The peak for Pan_tro_3.0 is shifted to higher values by an order of magnitude. (B) Increase in contig N50 for all chromosomes that were not finished with clones in Pan_tro_2.1.4 or Pan_tro_3.0. (C) Length distribution of filled gaps in Pan_tro_3. Negative values constitute wrongly separated overlapping contig ends in Pan_tro_2.1.4. (D) Increase in annotated interspersed repeats separated by repeat family.