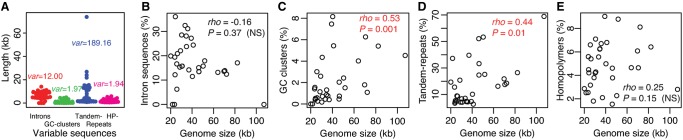
Fig. 2.
—Contribution of introns, GC-clusters, tandem-, and homopolymer-repeats to mitogenome variation among 33 yeast species. (A) Beeswarm plot (with variances) of sequence lengths of introns, GC-clusters, tandem-, and homopolymer (HP)-repeats. (B–E) Scatter plots (with Spearman’s correlation coefficient ρ and P value) of mitogenome size against the proportion of (B) introns, (C) GC-clusters, (D) tandem-, and (E) homopolymer-repeats in each mitogenome. To ease readability, significantly positive correlations are colored in red, whereas nonsignificant (NS) correlations are in black.