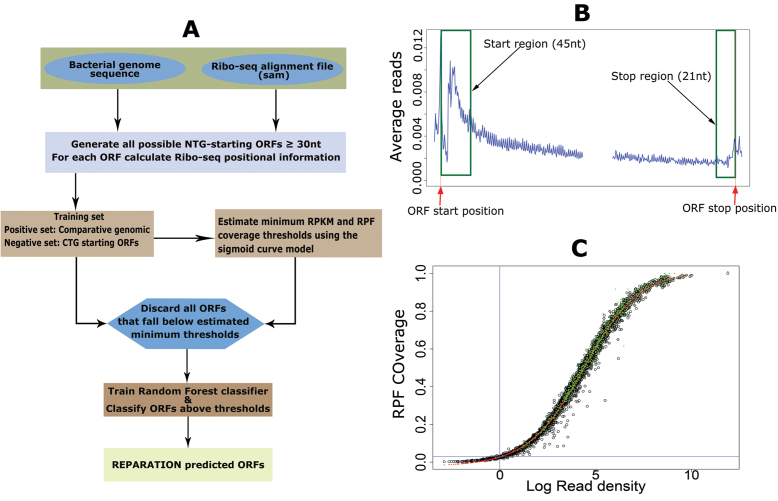
Figure 1.
RibosomeE Profiling Assisted (Re-)AnnotaTION (REPARATION) pipeline for de novo open reading frame (ORF) delineation in prokaryotes. (A) REPARATION workflow diagram. The entire prokaryotic genome is traversed and all possible NTG-starting ORFs are generated. Next, ORF-specific positional Ribo-seq signal information is calculated based on the metagene profile (B). To discard spurious ORFs, the minimum log RPKM and ORF ribosome protected mRNA footprint (RPF) coverage thresholds are estimated using a four-parameter logistic sigmoid curve (S-curve) (C). (B) Metagene profile of salmonella data indicating read accumulation at the start and stop of ORFs (stitched together in the middle for visualization purposes). (C) S-curve with fitted four parameters logistic curve (red) and indication of predicted ORFs with support from N-terminal proteomics data (green) in the case of Escherichia coli.