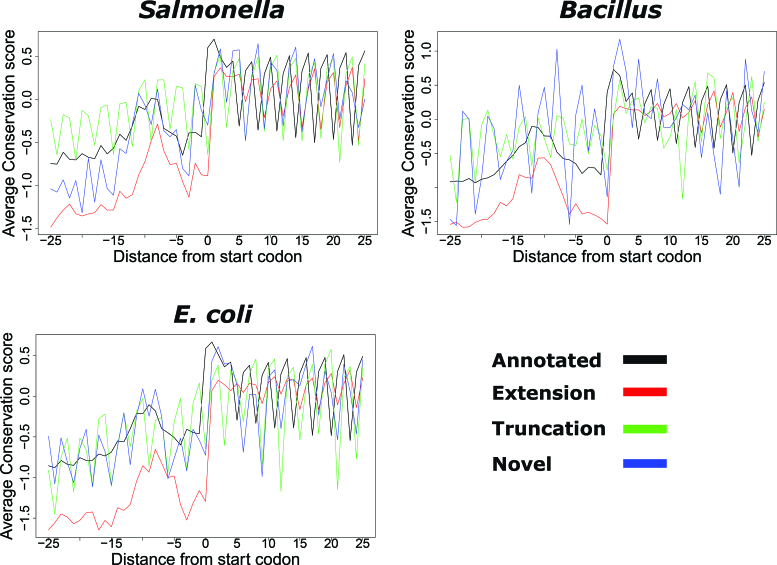
Figure 3.
Conservation pattern of REPARATION predicted ORFs. Nucleotide conservation scores are calculated using the Jukes cantor conservation matrix for nucleotides. Site conservation scores are calculated using the rate4site algorithm and displayed for a ±25 nt window around the predicted start site. The site conservation score was calculated only for ORFs with at least five orthologous sequences from a collection of randomly selected bacteria protein sequences from species within the same family as Salmonella/Escherichia coli and Bacillus and outside the family. A total of 833 annotated, 161 extensions, 99 truncations and 12 novel ORFs had at least five orthologous sequences in case of Salmonella, while the E. coli profile consisted of 2359 annotated ORFs, 70 extensions, 26 truncations and 18 novel ORFs. In the case of Bacillus, 1886 annotated, 112 extensions, 19 truncations and 2 novel ORFs were considered.