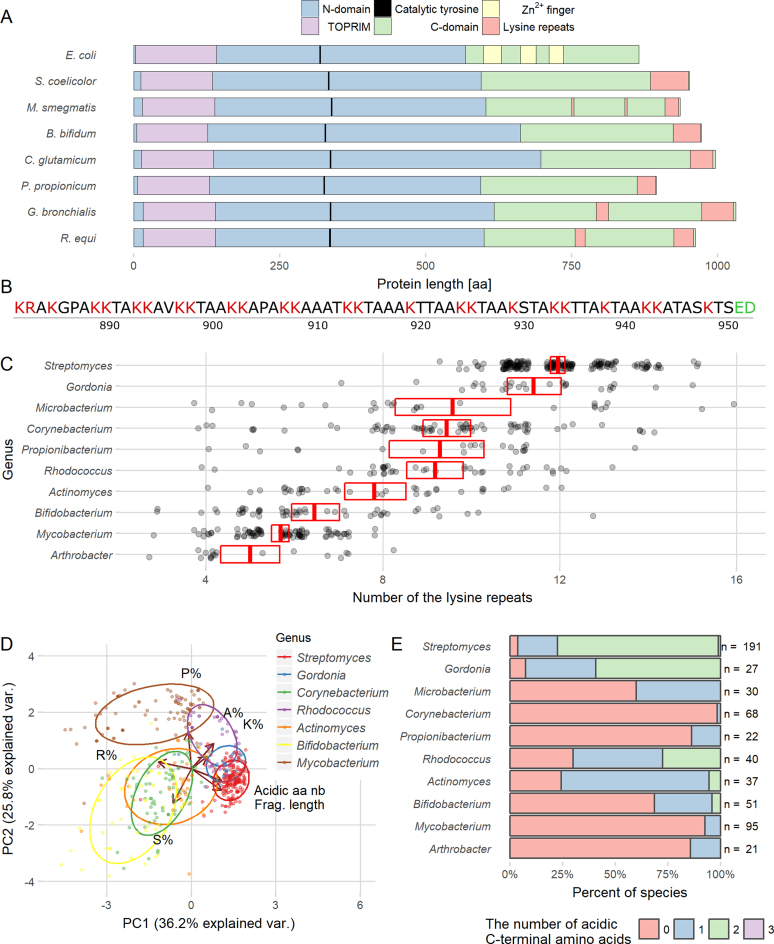
Figure 1.
Lysine repeats are characteristic of the actinobacterial TopA C-terminal domain. (A) Comparison of TopA sequences from various bacterial species. Blue and green color bars represent N- and C-terminal domains, respectively. Positions of the TOPRIM domain (purple), the catalytical tyrosine residue (black), Zn2+ fingers (yellow) and the lysine repeats (red) are indicated. (B) Sequence of S. coelicolor TopA 71 terminal amino acids encompassing the lysine repeats (K1–2(A/T/N)3–5). Basic amino acids are marked in red, and acidic in green. (C) The number of lysine repeats present in the TopA C-terminal domains of various genera belonging to Actinobacteria. Only data for the genera that had at least 20 identified proteins (699 proteins in total) are presented. The red box with a crossbar indicates the mean with 95% confidence interval, and all observations are marked by semitransparent points. (D) Principal component analysis (PCA) of the lysine repeats found in TopA C-terminal domains of Actinobacteria showing the variation in length and amino acid content. PCA was performed using the amino acid composition shown as the percentage of each amino acid: proline (P%), lysine (K%), alanine (A%), arginine (R%) and serine (S%), the length of the lysine repeats (Frag. length) and the number of acidic amino acids present among the last three amino acids of the protein (Acidic aa nb). On the plot, the first two principal components that have the largest variance (indicated on the plot) are shown. Color points indicate individual species belonging to different genera with normal data ellipse for each group (probability = 0.68). The analyzed genera are indicated. (E) Percentage of Actinobacteria TopA homologs containing acidic amino acids at the C-terminus. The analyzed genera are indicated.