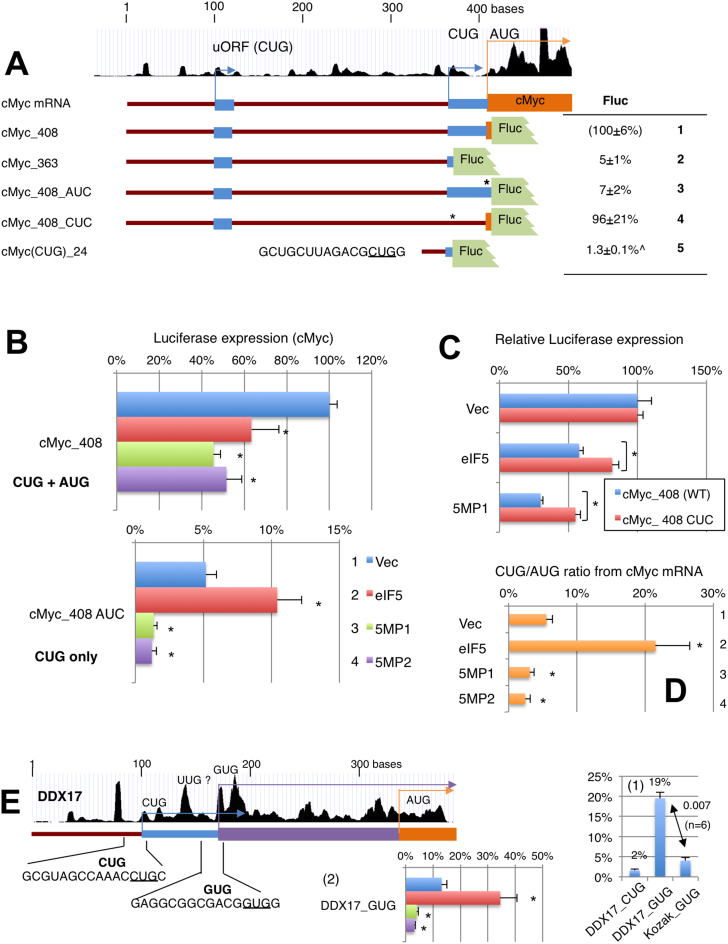
Figure 6.
Translational control of cMYC and DDX17 by eIF5 and 5MP. (A) Analysis of cMYC start codons. Top, schematics depict the cMYC leader region with UCSC ribosome profile and boxes representing reading frames color-coded as in Figure 3A. The table describes firefly luciferase activities from constructs listed to the left and depicted in the middle, relative to the activity from cMyc_408 (row 1). ∧, activity for cMyc(CUG)_24 is presented relative to that from Kozak AUG. Asterisks, start codon mutations. (B) Effect of eIF5 and 5MP1/2 expression was tested with indicated cMyc reporter plasmids, and presented as values relative to cMyc_408 vector control, as in Figure 2C. P values of significance from top; cMyc_408, 0.04, 0.00004 (n = 8), 0.01 (n = 4); cMyc_408 AUC, 0.01, 0.0007, 0.0005 (n = 10) compared to vector control. (C) Effect of eIF5 and 5MP1 on cMyc_408 and cMyc_408_CUC was compared to the values from vector control for each, and statistical significance was computed using students’ Ttest. *P values of significance; from top, 0.04, 0.02 (n = 4). (D) CUG/AUG ratio (R) in cMyc_408 (WT) was computed based on the following formula and presented with asterisks denoting statistical significance (P < 0.05) compared to the value with vector control. R = a/(a – b), where a is activity from cMyc_408 AUC and b is activity from cMyc_408 (WT). *P values of significance; from top, 0.01, 0.02, 0.009 (n = 10) compared to vector control. (E) DDX17 mRNA structure was depicted as in Figure 5A–C. Graph 1 indicates firefly luciferase activities from the reporter with DDX17 CUG and GUG codons carrying the 24-nt mRNA sequences preceding each (columns 1 and 2) and compared to activity from Kozak GUG (column 3). Graph 2, the effect of eIF5 and 5MP1/2 on the DDX17 GUG reporter translation. p values from top; *0.02, 0.003, 0.003 (n = 6) compared to vector control.