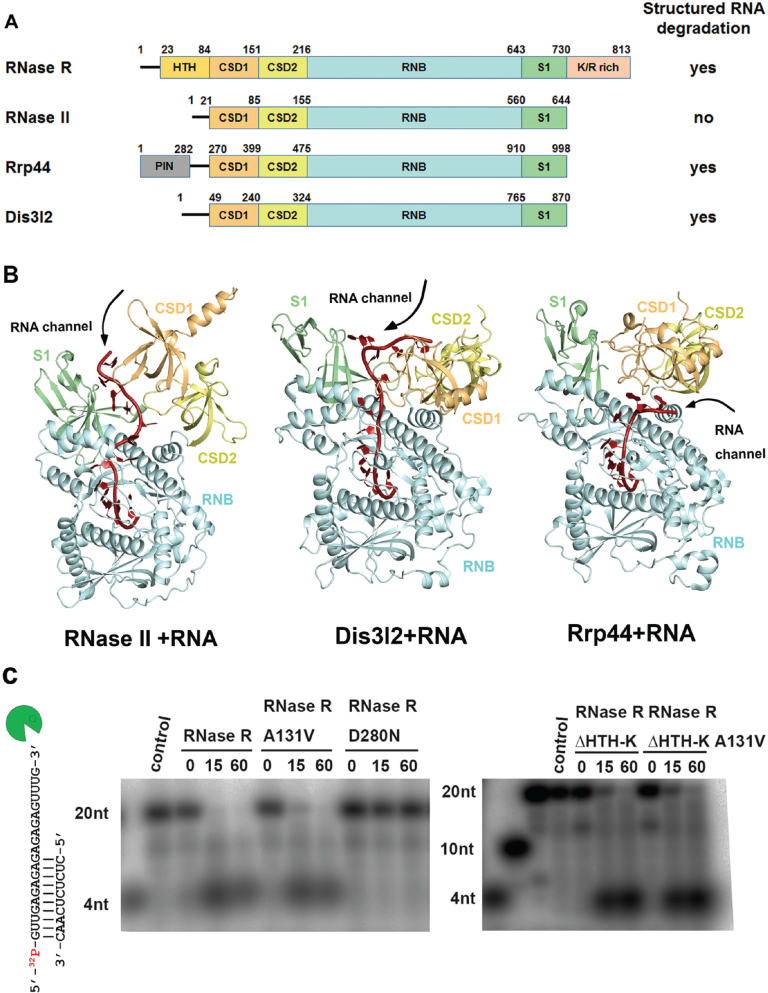
Figure 1.
Domain organization and structure comparison of the RNase II family of exoribonucleases. (A) Overall domain organization of the RNase II family proteins: RNase R, RNase II, Rrp44 and Dis3l2. RNase R, Rrp44 and Dis3l2 can degrade RNA with secondary structures, but RNase II cannot, as indicated at right. (B) Crystal structures of the RNA-bound form of RNase II (PDB: 2IX1), Dis3l2 (PDBID: 4PMW) and Rrp44 (PDB: 2VNU). The open top and side channels are marked on the structures. (C) Wild-type RNase R and A131V mutant (100 nM) unwound and degraded the 5′-end-32P-labeled dsRNA (2.5 nM), whereas the D280N mutant could not degrade dsRNA with a 3′ overhang (left panel). The truncated mutant RNase R ΔHTH-K and its A131V mutant (crystallized protein in this study) retained their activity in dsRNA unwinding and degradation (right panel).