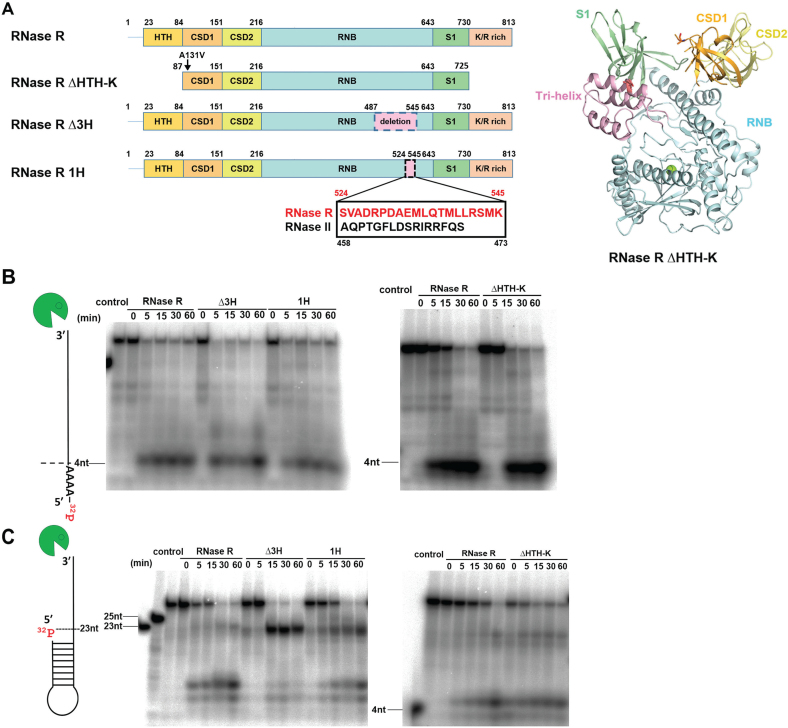
Figure 3.
A tri-helix ‘wedge’ region in the RNB domain of RNase R is crucial for its RNA unwinding activity. (A) Two mutants were constructed; a tri-helix region (residues 487–545) was deleted from the tri-helix mutant RNase R Δ3H, whereas a 21-residue helix (residues 524–545) was replaced by the corresponding helix from RNase II (residues 458–473) in the RNase R 1H mutant. The tri-helix region is colored in pink in the crystal structure of RNase R ΔHTH-K displayed in the right panel. (B) RNA degradation assays show that RNase R and its mutants Δ3H, 1H and ΔHTH-K (100 nM) fully degraded the 40-nt single-stranded RNA (2.5 nM) to a final product of ∼4 nt. (C) Full-length RNase R and truncation mutant ΔHTH-K degraded the stem–loop RNA with a 3′ overhang to a final product of ∼4 nt, but the wedge mutants Δ3H and 1H could not fully degrade the stem–loop RNA and generated major digested products of ∼23 nt.