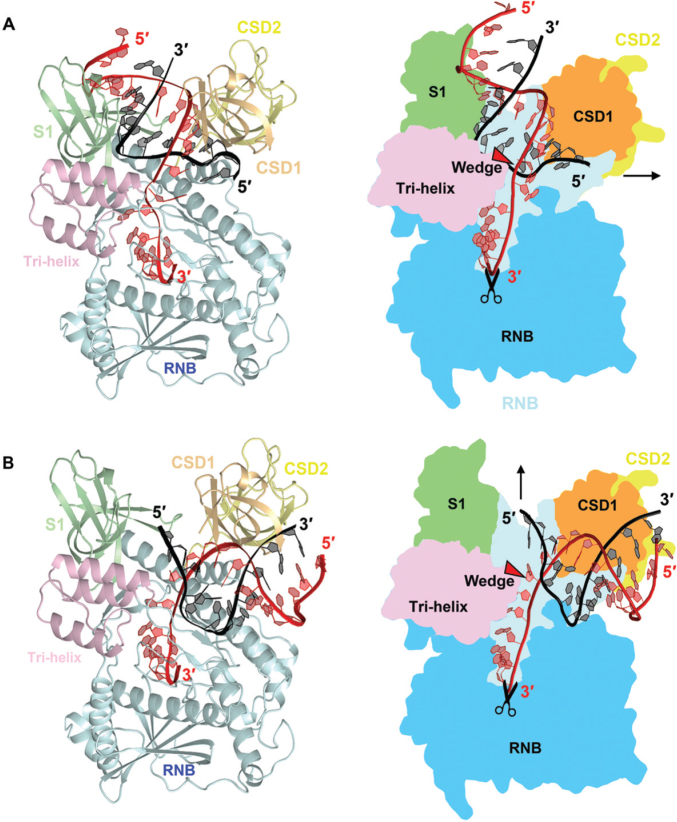
Figure 5.
Working models for RNA binding, unwinding and degradation by RNase R. (A) An RNase R-RNA model was built based on the crystal structure of RNase II bound with RNA (PDB: 2IX1). The 3′ overhang of duplex RNA enters all the way into the active site from the top channel. The RNA duplex region is bound by the CSD1 and S1 domains in the top channel and, upon reaching the tri-helix wedge region in the RNB domain, the RNA duplex is unwound. (B) A second RNase R-RNA model was built based on the crystal structure of Rrp44 bound with RNA (PDB: 2VNU). The 3′ overhang of duplex RNA is guided through the side channel into the active site in the RNB domain. The RNA duplex region is bound by the CSD1 and RNB domains in the side channel and, upon reaching the tri-helix wedge region, the RNA is unwound. It should be noted that the side channel in RNase R is not wide enough to accommodate a duplex RNA, however, the CSD1/2 domains may be flexible and change their conformation upon RNA binding.