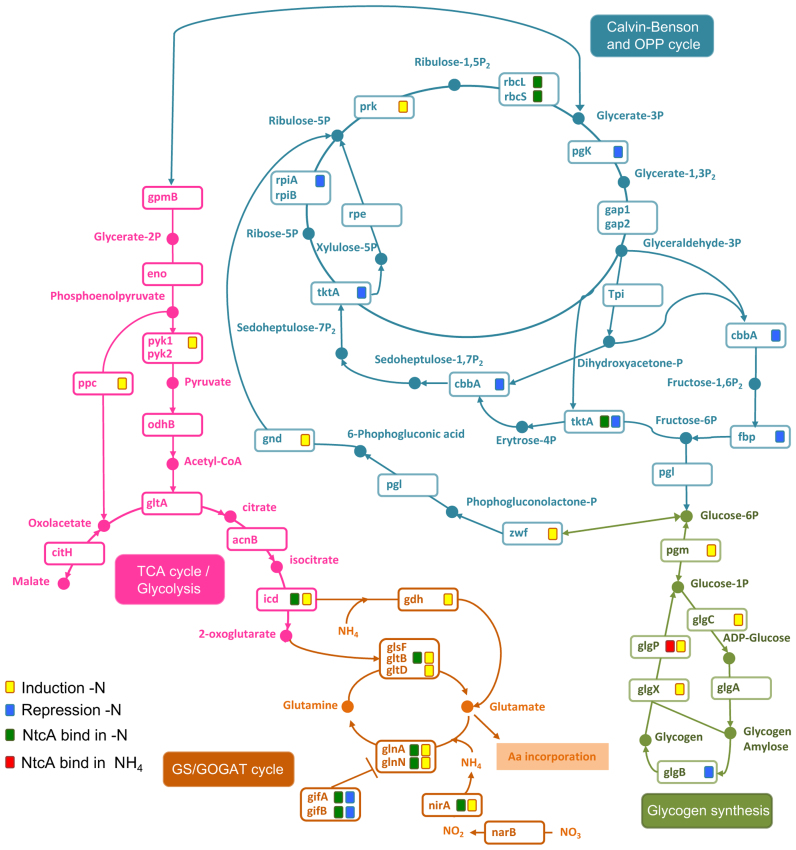
Figure 3.
Changes in expression of genes involved in nitrogen assimilation and carbon metabolism. This diagram shows metabolic pathways and metabolites of the TCA and GS/glutamine oxoglutarate amidotransferase (GOGAT) cycle, glycolysis, gluconeogenesis, pentose phosphate pathway (OPP) and glycogen metabolism. These pathways were predicted from the KEGG database (www.genome.jp/kegg/pathway.html). NtcA target genes obtained from ChIP-seq analysis are shown for −N (green square) or NH4 (red square) treatments. Genes encoding metabolic enzymes are shown below; narB, nitrate reductase; nirA, nitrite reductase, gifA, GS-inactivating factor; gifB, GS-inactivating factor; glnA, GS type I; glnN, GS type III; gltD, glutamate synthase small subunit; gltB, GOGAT; glsF, ferredoxin-dependent glutamate synthase, gdhA, glutamate dehydrogenase; icd, isocitrate dehydrogenase; acnB, aconitate hydratase; citH, malate dehydrogenase; gltA, citrate synthase; odhB, pyruvate dehydrogenase; pyk1–2, pyruvate kinase; eno, enolase; ppc, phosphoenolpyruvate (PEP) carboxylase; gpmB, phosphoglycerate mutase; pgk phosphoglycerate kinase; gap1, glyceraldehyde-3-phosphate dehydrogenase (catabolic reaction); gap2, glyceraldehyde-3-phosphate dehydrogenase (anabolic reaction); tpi, glycogen isoamylase; cbbA, fructose-bisphosphate aldolase; fbp, fructose-1,6-bisphosphatase; pgl, 6-phosphogluconolactonase; pgm, phosphoglucomutase; glgC glucose-1-phosphate adenylyltransferase; glgP, glycogen phosphorylase; glgX, glycogen isoamylase; glgA, glycogen synthase; glgB, 1,4-alpha-glucan branching enzyme; zwf, glucose-6-phosphate dehydrogenase; gnd, 6-phosphogluconate dehydrogenase; tktA, transketolase; rpiA or B, ribose-5-phosphate isomerase; rpe, pentose-5-phosphate-3-epimerase; prk, phosphoribulokinase; rbcL, ribulose bisphosphate carboxylase large subunit; rbcS, ribulose bisphosphate carboxylase small subunit.