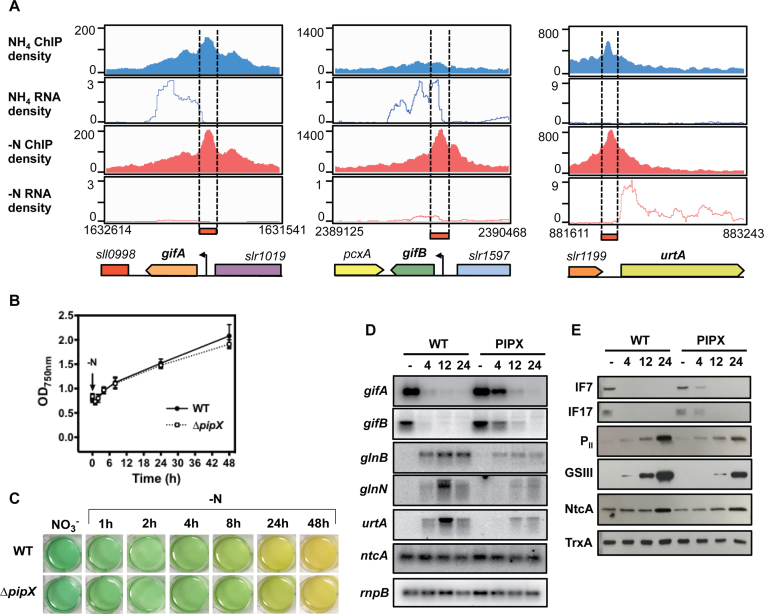
Figure 9.
The role of PipX in the NtcA-regulated genes. (A) Read coverage of NtcA-binding peaks assigned to gifA, gifB and urtA. Their ChIP-seq and RNA-seq density profiles are shown in blue for NH4 and in red for −N conditions. (B) Growth of Synechocystis WT and ΔpipX strains after nitrogen step-down. Cells of WT and ΔpipX strains cultivated to their midlog growth phase in BG11C medium supplemented with NH4 were transferred to nitrogen-free BG11C medium.Their growth was measured at OD750 for 48 h. (C) Images of cyanobacterial cell suspensions of Synechocystis WT and ΔpipX obtained during growth curve analysis (panel B) at indicated times. (D) Northern blot analysis of expression of gifA, gifB, glnB, glnN, urtA and ntcA in response to nitrogen starvation. Total RNA was isolated from WT cells transferred from BG11C supplemented with ammonium to nitrogen-free BG11C medium for 24 h. The filters were hybridized with gifA, gifB, glnB, glnN, urtA and ntcA probes and subsequently stripped and rehybridized with rnpB probe as a control. (E) Western blot analysis of IF7, IF17, PII, GSIII and NtcA in response to nitrogen starvation. Cells were grown in BG11C supplemented with ammonium to midlog growth phase and then transferred to nitrogen-free BG11C medium and cultivated for 24 h. Samples of 10 μg of total proteins from soluble extracts were separated by 15% SDS/PAGE and analyzed by western blots to detect IF7, IF17, PII, GSIII, NtcA and TrxA.