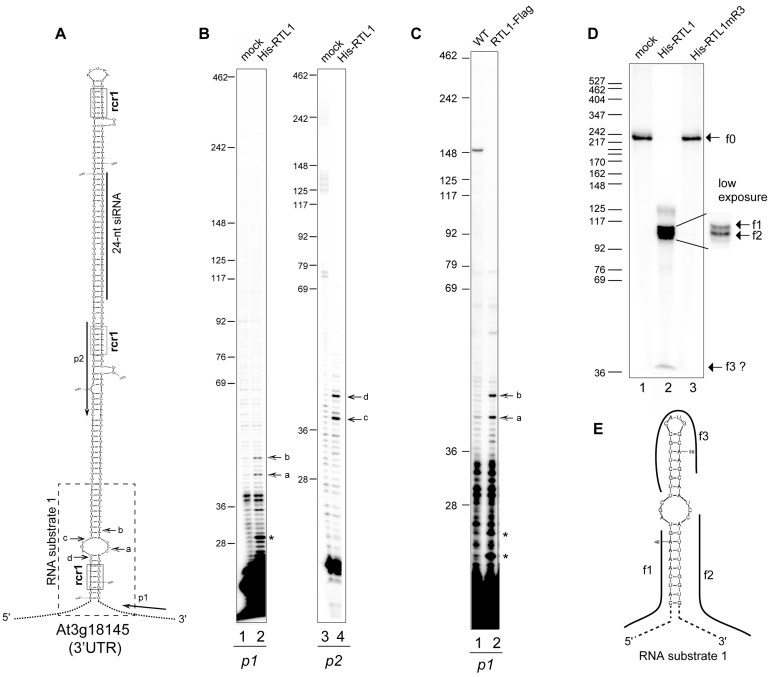
Figure 1.
RTL1 site-specifically cleaves a near-perfect duplex structure in vitro and in planta. (A) Left, the hairpin structure predicted from 3′UTR At3g18145 mRNA sequence is shown. The positions of p1 and p2 primers used in primer extension experiments are indicated. The dashed rectangle indicates the sequence corresponding to the RNA substrate-1 shown in (E). The sequence corresponding to a 24-nt small RNA is shown by a bar. Arrows show cleavage sites a–d mapped by primer extension. The RNA duplex consensus region rcr1 are boxed (B) Primer extension analysis using in vitro transcribed 3′UTR -At3g18145 and p1 or p2 primers. Arrows (a–d) show mapped cleavage sites in both strands. (C) Primer extension analysis on total RNA extracted from Col0 and overexpressing RTL1-Flag #1 plants with p1 primer. Arrows show the rcr1 cleavage positions. (D) Cleavage assay of 32P-CTP RNA substrate-1 using His-RTL1 or His-RTL1m3 proteins. Arrows indicate non cleaved RNA substrate-1 (f0) and cleavage fragment products (f1–f3). A low exposure insert is shown to better visualize the f1 and f2 fragments. DNA size markers are indicated on the left. (E) The predicted 32P-CTP RNA substrate-1 structure and f1–f3 products are represented.