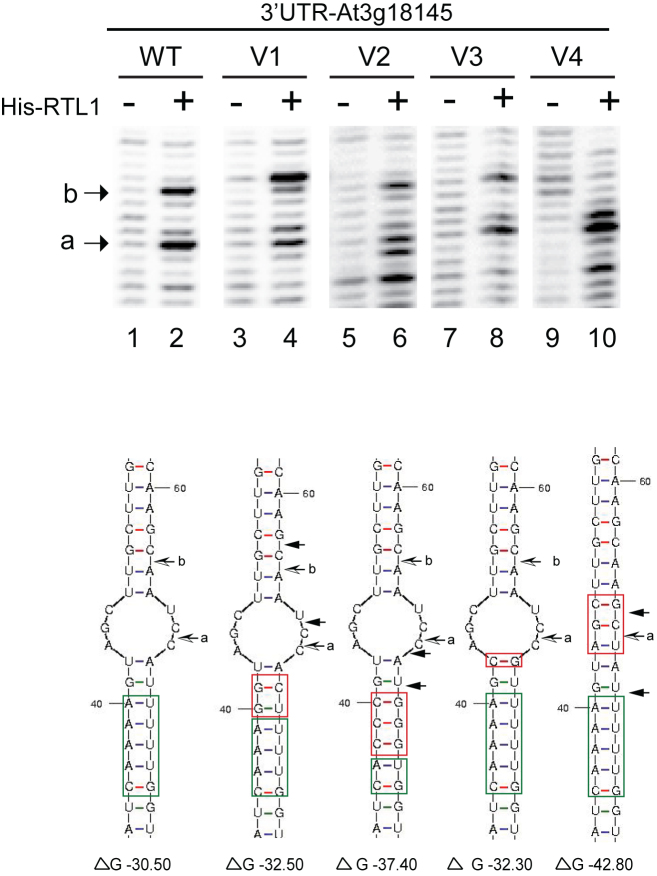
Figure 2.
RNA sequence and structure requirements for RTL1 cleavage. Primer extension analysis using in vitro transcribed 3′UTR RNA (WT and mutated V1-V4) incubated with His-RTL1 protein (+) or buffer alone (−). In the version V1 the A::U and the G::U base pairs within and next to the RNA duplex motif were mutated to G::U and G::C respectively; in V2, three of the A::U base within the RNA duplex motif were mutated to C::G; in V3 the U:: A base pair located in the loop structure was mutated to C::G and in the -V4, the UCG nucleotides in loop structure were mutated to GCU. The Wt and mutated sequences are green and red boxed respectively. Arrow shows cleavage site mapped with p1 primer. Note the two novel cleavage sites (black arrows) adjacent to the a and b sites (black and white arrows) in V1 and the novel cleavage sites adjacent to the a site in V2 and V4. The ΔG (kcal/mol) determined by RNA Mfold for each RNA substrate is indicated.