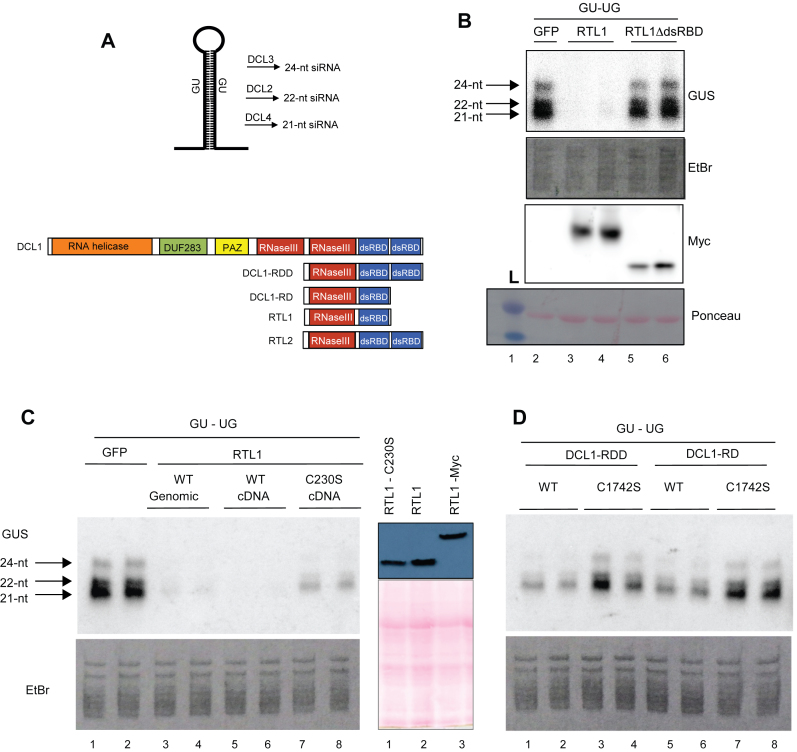
Figure 4.
The native dsRBD and its conserved cysteine C230 are required for RTL1 cleavage of RNA in planta. (A) Top, schematic representation of the GU-UG RNA and of the siRNAs produced from the processing of the dsRNA stem by DCL2, DCL3 and DCL4. The GU-UG siRNA construct contains promoter (p35) and terminator (t35S) sequences derived from 35S Cauliflower mosaic virus (CaMV) Bottom, schematic representation of native DCL1, RTL1 and RTL2 and of the truncated proteins DCL1-RD and DCL1-RDD mimicking RTL1 and RTL2, respectively. (B) Nicotiana benthamiana leaves co-infiltrated with a 35S:GU-UG construct (GU-UG) and either a control 35S-driven construct (GFP), a WT-tagged 35S:RTL1-Myc construct (RTL1) or a mutant 35S:RTL1ΔdsRBD-Myc construct lacking the dsRBD (RTL1ΔdsRBD). Low molecular weight RNAs were extracted and hybridized with a GU probe. Ethidium bromide-stained RNAs are shown as loading control. Proteins were extracted and hybridized with a Myc antibody. Ponceau staining (pink dye) is shown as protein loading control (lanes 2–6). L, indicates protein ladder. (C). Left, N. benthamiana leaves co-infiltrated with a 35S:GU-UG construct and either a control 35S-driven construct (GFP), a genomic-based or cDNA-based WT 35S:RTL1 construct (WT genomic and WT cDNA), 35S:RTL1 constructs mutated at cysteine 230 (C230S). Right, western blot analysis show comparable expression of 35S:RTL1 C230), 35S:RTL1 and 35S:RTL1-Myc constructs in N. benthamiana leaves. Proteins were extracted and hybridized with α RTL1 antibody. Ponceau staining (pink dye) is shown as protein loading control (lanes 1–3). (D) Nicotiana benthamiana leaves co-infiltrated with a 35S:GU-UG construct and either a control 35S-driven construct (GFP), a genomic-based or cDNA-based WT 35S:RTL1 construct (WT genomic and WT cDNA), 35S:RTL1 constructs mutated at cysteine 230 (C230S) and truncated 35S:DCL1 constructs (DCL1-RDD and DCL1-RD) carrying WT DCL1 sequence (WT) or a mutation at cysteine 1742 (C1742S). Note that RNA samples shown in (C) and d were run on a unique gel, so that the control 35S:GU-UG + 35S:GFP for (D) is visible on (C).