Figure 8.
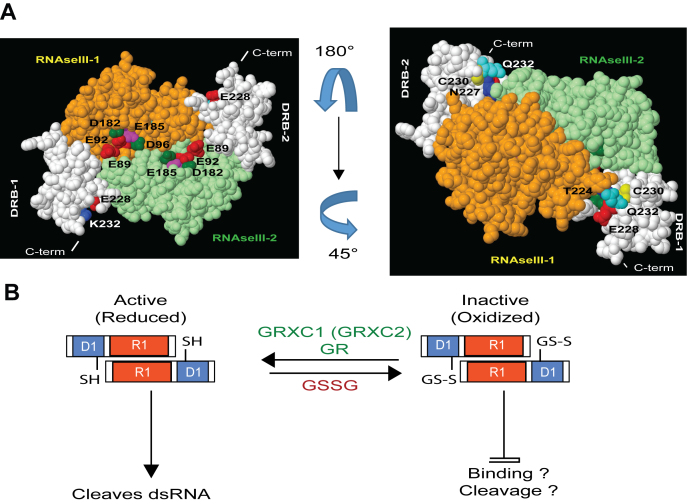
In silico and functional analysis of RTL1 suggest a novel regulatory mechanism for RNase III activity in plants. (A) Modeled RTL1 (residues 50–284) homodimer based on mouse Dicer (3c4b.1.A) (23). The RNase III domain of two RTL1 molecules are shown in orange and green while both dsRBD are shown in white. In the left panel, the residues E89, E92, D96 and E185 (E37, E40, D44 and E110 in Aquifex aeolicus RNase III) located in the RNase III domain and required for RNA cleavage are shown. In the right panel, the RTL1 homodimer was rotated 180° and 45° to show conserved cysteines C230 in each dsRBD. The residues T224, N227, E228 and Q231 (T154, Q15, E158 and Q161 in A. aeolicus RNase III) located near to the C230 are indicated. These residues are required for RNA binding of Aa-RNase III. (B) In the proposed model, the cysteine C230, which is essential for cleavage activity, is kept in its reduced state (-SH). Glutathionylation of C230 (S-SG) in RTL1 sequence does not affect RTL1 dimerization but it might inhibit dsRNA binding and/or cleavage activity. RTL1 activity inhibition is reversible upon treatment with GRXC1 or GRXC2.