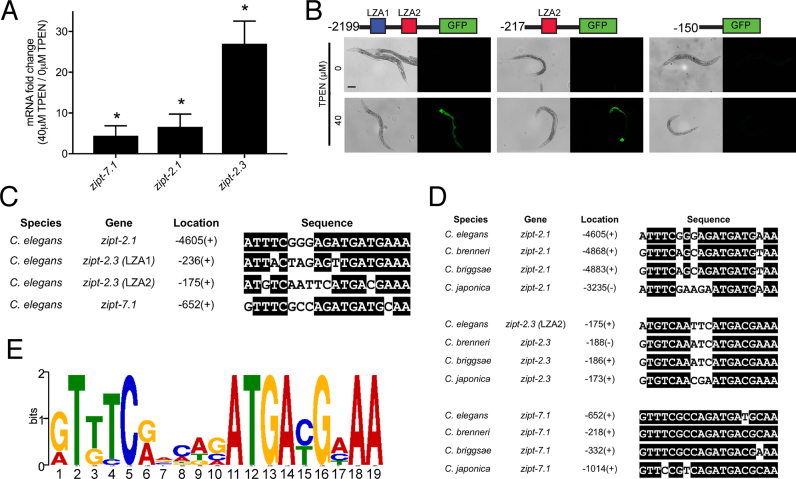
Figure 1.
Identification of the LZA element. (A) A population of mixed-stage, wild-type animals were cultured with 0 or 40 μM TPEN for 16 h. RNA was analyzed by qRT-PCR. Values are the ratio of mRNA levels at 40 μM TPEN/0 μM TPEN, an indication of transcriptional activation and are the average of three biological replicates; error bars display standard deviation. Each gene displayed significantly increased mRNA levels at 40 μM TPEN (*P < 0.05). (B) Diagrams of the zipt-2.3 promoter region (not to scale) extending 2199, 217 or 150 bp upstream of the translation start site fused to the GFP coding region. Boxes show LZA1 element (blue) and LZA2 element (red). Transgenic animals containing these constructs were cultured with 0 or 40 μM TPEN for 16 h and imaged with bright field to display worms (left) or fluorescence to display GFP expression (right). Scale bar indicates 100 μm. (C) An alignment of LZA elements from the promoters of Caenorhabditis elegans zipt-2.1, zipt-2.3 and zipt-7.1. Identical residues are highlighted in black. Location is the position of the first base pair of the LZA element relative to the ATG start codon, and +/− refers to the orientation relative to the DNA strand that is transcribed. (D) Sequence alignments of LZA elements in the promoter regions of zipt-2.1, zipt-2.3 and zipt-7.1 in the Caenorhabditis species elegans, brenneri, briggsae and japonica identified by MEME. For zipt-2.3, the LZA2 element in C. elegans is conserved in three other species, and the LZA1 element is not conserved. (E) A position weight matrix of the LZA element based on 12 motifs shown in D. The height of each nucleotide represents the frequency scaled in bits.