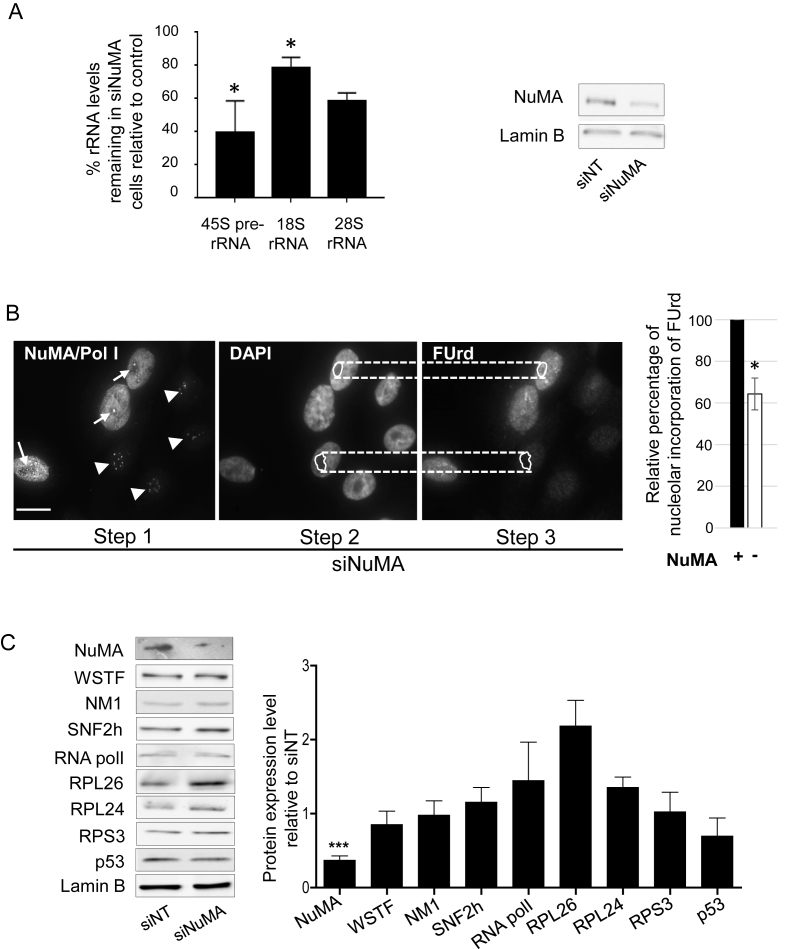
Figure 4.
Silencing NuMA reduces rRNA levels with no significant impact on proteins associated with ribosomal biogenesis or p53-dependent nucleolar stress response. S1 cells were transfected with siRNA for NuMA (siNuMA) or non-targeting sequence (siNT) and left in culture until day 8. (A) Quantitative RT-PCR for 45S prerRNA, and related 18S RNA and 28S RNA. Representative western blot of the decrease in NuMA expression upon siRNA treatment is shown. Lamin B is used as loading control. (B) FUrd labeled nascent rRNA in siNuMA treated cells. Analysis was done by triple immunostaining for NuMA, Polymerase I [Pol I] and FUrd. The unique location of Pol I in the nucleolus (Step 1) was used to confirm that the weaker areas on DAPI stained nuclei are indeed nucleoli (Step 2). Upon delineating the nucleolar area on DAPI images, this area was then used for corresponding FUrd stained nuclei (Step 3) to perform the analysis as indicated in the ‘Materials and Methods’ section. Cells silenced for NuMA are revealed by the absence of immunostaining in the nucleoplasm (see nuclei with arrowhead indicating Pol I staining; nuclei that still show NuMA staining include an arrow to indicate Pol I staining). Bar graph shows the relative intensity of FUrd staining in the nucleoli of cells with (+) or without (−) NuMA expression. Over 30 cells were analyzed for each treatment condition (n = 3). (C) Protein quantification by western blot analysis in cells treated with siNuMA relative to levels in siNT-treated cells. Shown are representative western blot images. Lamin B is used as loading control. Size bar, 10 μm; *P < 0.05, ***P < 0,001; one sample t-test.