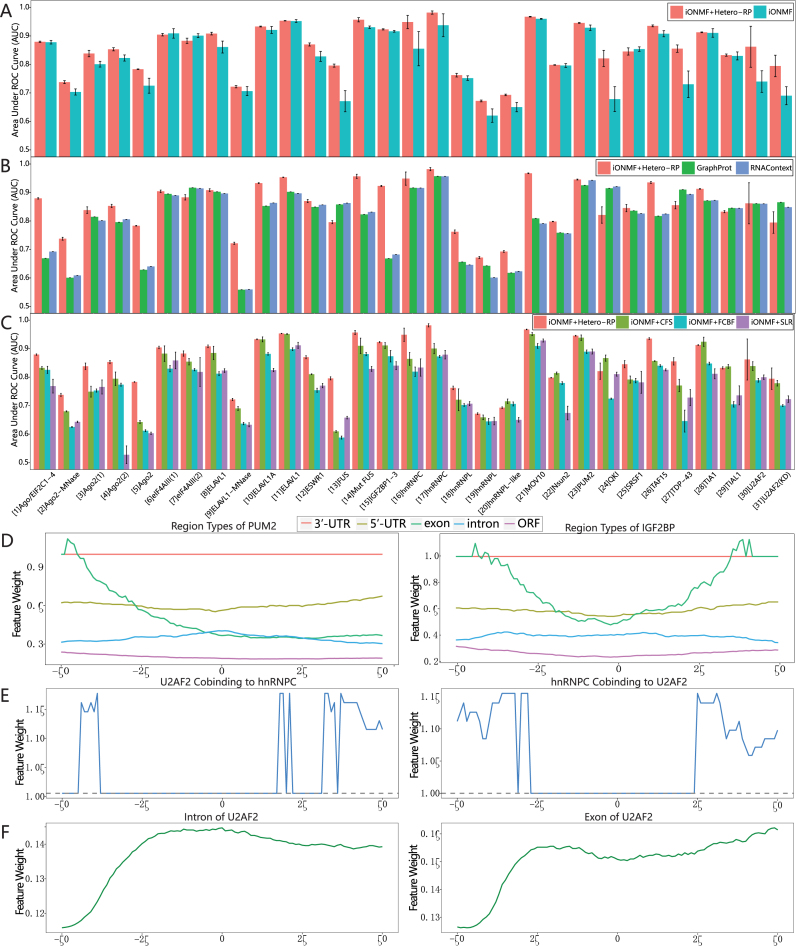
Figure 3.
Incorporating Hetero-RP in RBP binding sites prediction. (A) iONMF with or without Hetero-RP is applied to 31 published CLIP experiments. The positive-links and negative-links sets are constructed according to the labels of the nucleotide positions in the training set. The performance is evaluated by the area under the receiver operating characteristic curve (AUC). (B) Hetero-RP is compared against state-of-the-art methods. (C) Hetero-RP is compared against popular feature selection methods. (D–F) Interpretations of the scales obtained by Hetero-RP. (D) The 3′-UTR region type of PUM2 and IGF2BP has the largest weights, consistent with the fact that the binding sites of both IGF2BP and PUM2 are distributed across 3′-UTRs. (E) The mutual co-binding of hnRNPC and U2AF2 has large weights and this observation agrees with the fact that hnRNPC interacts with the same positions as U2AF2. Moreover, the weights are even larger at the upstream and downstream, supporting the evidence of direct competition between the two. (F) The intron and exon region types of U2AF2 start to scale up at ∼30 nucleotides upstream from the binding site, where the reported intron–exon boundary is located.