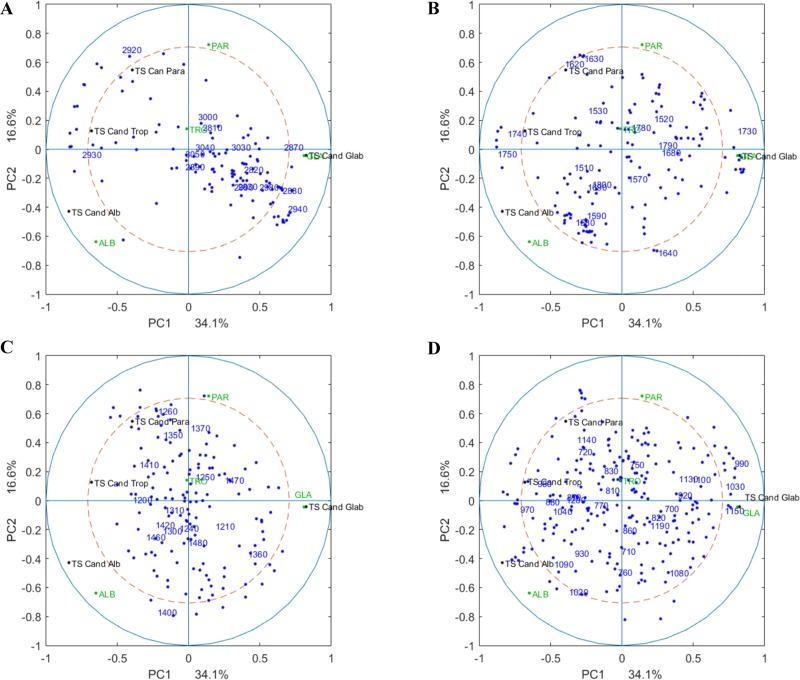
Fig 2. Correlation loading plot (PC1 and PC2) of NGS and FT-IR (block 1, 2, 3 and 4) with global scores.
The correlation loading plots showing the correlation between the global scores of the CPCA analysis with the four different FT-IR blocks and the distance matrix of the genetic data. In addition, the correlations between the global scores and the genetic distance matrix and the indicator variables for each species are visualized in each plot. (A) Correlations between global scores and the lipid region (block 1, 3050–2800 cm-1), the genetic distance matrix and the species indicator variables; (B) correlations between the global scores and the mixed lipid and protein region (block 2, 1800–1500 cm-1), the genetic distance matrix and the species indicator variables; (C) correlations between the global scores and the mixed lipid, protein and polysaccharide region (1500–1200 cm-1), the genetic distance matrix and the species indicator variables and (D) correlations between the global scores and the polysaccharide region (1200–700 cm-1), the genetic distance matrix and the species indicator variables. Blue dots represent FT-IR wavelengths; black dots distances to TS Cand alb, TS Cand para, TS Cand glab and TS Cand trop represent type strains (TS) of the four species and green dots namely ALB, PAR, GLA and TRO represent the group variables (indicator variables).