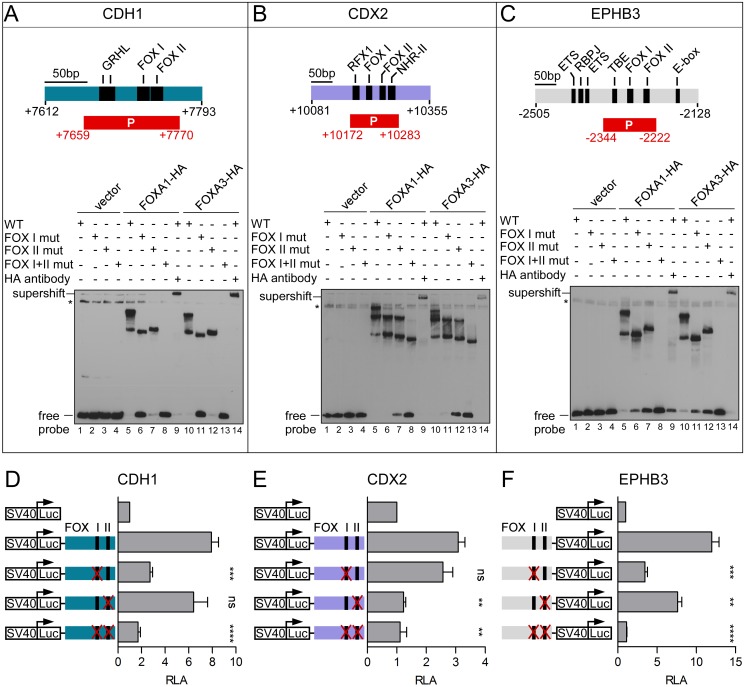
Fig 6. CDH1, CDX2 and EPHB3 enhancer activity is dependent on FOXA binding.
(A) (Top) Scheme of the ECR at +7.8 kb at the CDH1 locus with the described tandem GRHL binding sites [42] and two potential FOX binding motifs. (Bottom) EMSA to test for in vitro binding of FOXA1 and FOXA3 to the CDH1 +7.8 kb enhancer. Asterisks: non-specific bands. mut: mutated, P: probe. (B) (Top) Scheme of the CDX2 +10.0 kb enhancer with the described RFX1, FOXA and nuclear hormone receptor-II (NHR-II) transcription factor binding sites [47]. (Bottom) EMSA to test for in vitro binding of FOXA1 and FOXA3 to the CDX2 +10.0 kb enhancer. (C) (Top) Scheme of the EPHB3 −2.3 kb enhancer with its known transcription factor binding sites. TBE: TCF/LEF-binding element. (Bottom) EMSA to test for in vitro binding of FOXA1 and FOXA3 to the EPHB3 −2.3 kb enhancer. (D, E, F) Luciferase reporter assay in LS174T cells with constructs covering the CDH1 (D), the CDX2 (E), and the EPHB3 (F) enhancer. Mutations of the respective FOX binding motifs are indicated by red crosses. Shown is the mean and SEM; n≥3. RLA: relative luciferase activity. Statistical significance was calculated relative to the wild-type luciferase reporter constructs.