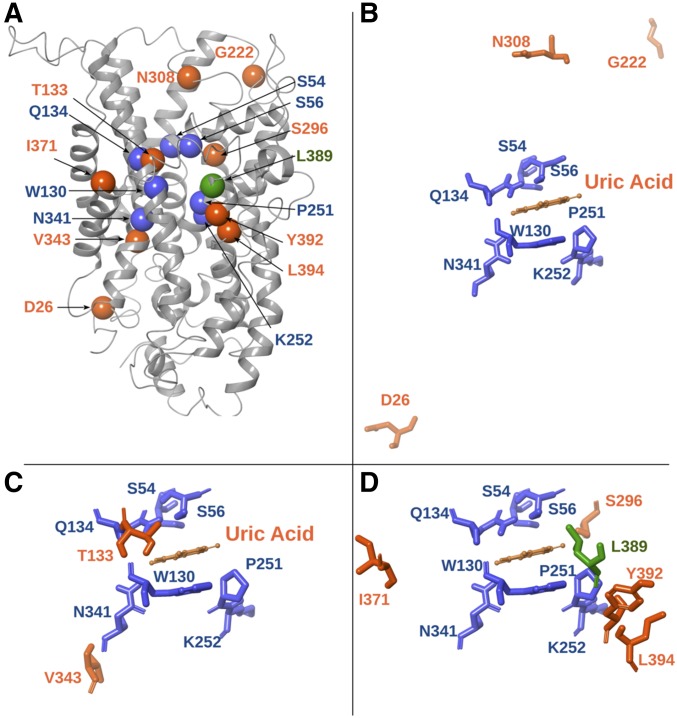
Figure 5.
Location of suppressor mutations in the modeled FurE structure. (A) Model structure of FurE highlighting the residues altered in suppressor mutations (orange spheres) in relation to the substrate-binding site residues (blue spheres), and a critical residue in the putative outer gate (green sphere) (Krypotou et al. 2015). (B) Topology of amino acids (Asp26 in LN, Asn308 in L7, and Gly222 in L5) modified in type III suppressors, located in loops that may function as flexible hinges (see text). (C) Topology of amino acids [Thr133 in transmembrane segment (TMS)3 and Val343 in TMS8] modified in type I suppressors (see text), located proximal to residues shown to interact with substrates in the major substrate-binding pocket of FurE (Ser54 and Ser56 in TMS1, Gln134 and Trp130 in TMS3, Pro251 and Lys252 in TMS6, and Asn342 in TMS8). (D) Topology of amino acids (Ser296 in TMS7, Ile371 in TMS9, and Tyr392 and Leu394 in TMS10) modified in type II suppressors, located next to or in the periphery of a major residue of the outer gate (Leu389 in TMS10).