Fig. 3.
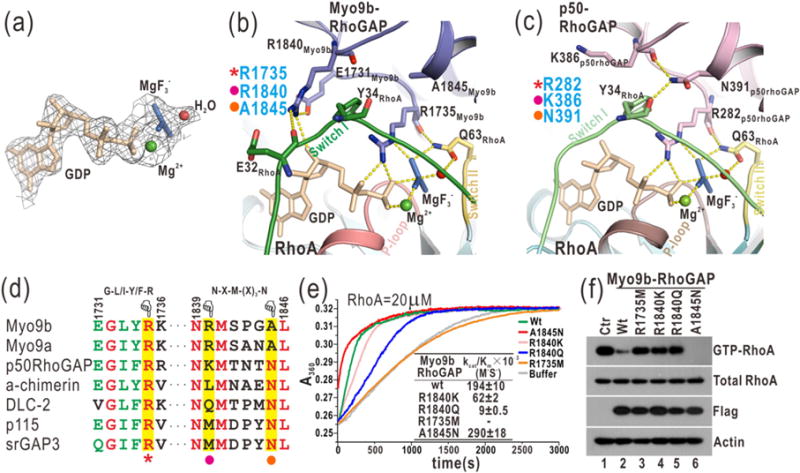
Myo9b-RhoGAP contains two arginine fingers for RhoA GTP hydrolysis. (a) A close-up view of GDP and in the complex structure by a stick model representation. The electron density map (2Fo-Fc map) of GDP, , Mg2+, and a putative nucleophilic water molecule is shown and contoured at 1.5 σ level. (b) A combined ribbon-and-stick model illustrates the catalytic site in the structure of the Myo9b-RhoGAP/RhoA complex. In this drawing, Myo9b-RhoGAP and the Switch I and II and P-loop of RhoA are colored in blue, green, yellow, and pink, respectively, and the residues essential for catalysis are shown as sticks. Hydrogen bonds and salt bridges are indicated by dashed lines. (c) A combined ribbon-and-stick model illustrates the catalytic site in the structure of the p50-RhoGAP/RhoA complex (PDB code: 1OW3). p50-RhoGAP is colored in violet. The color schemes of RhoA follow that shown in panel (b), and the residues essential for catalysis are shown as sticks. (d) Two selected key regions of the structure-based sequence alignment of the RhoGAP domains from different proteins. The identical and highly conserved residues are colored in red and green, respectively. The catalytic arginine finger, the auxiliary arginine finger, and the essential asparagine in the RhoGAP domains are highlighted with a red star, purple dot, and yellow dot, respectively, at the bottom. (e) Time courses of GTP hydrolysis for RhoA (20 μM) catalyzed by Myo9b-RhoGAP and its mutants (40 nM). The Michaelis–Menten kinetic parameters (kcat/Km) are summarized as an inset. Data shown are mean values ± SD from two independent experiments. (f) Biochemical pull-down analysis of active GTP-bound RhoA with Myo9b-RhoGAP and its mutants. The active RhoA levels were measured by GST pull-down analysis with GST-RBD and analyzed by Western blotting using a specific anti-RhoA antibody. The levels of total RhoA, Flag-tagged Myo9b-RhoGAP, and the actin (as an internal loading control) in the cell lysate were also shown. Consistently, mutations of the two arginine fingers impaired the catalytic activity of Myo9b-RhoGAP, while the A1845N mutation increased that of Myo9b-RhoGAP.
