Abstract
The increased threat of terrorism across the globe has raised fears that certain groups will acquire and use radioactive materials to inflict maximum damage. In the event that an improvised nuclear device (IND) is detonated, a potentially large population of victims will require assessment for radiation exposure. While photons will contribute to a major portion of the dose, neutrons may be responsible for the severity of the biologic effects and cellular responses. We investigated differences in response between these two radiation types by using metabolomics and lipidomics to identify biomarkers in urine and blood of wild-type C57BL/6 male mice. Identification of metabolites was based on a 1 Gy dose of radiation. Compared to X rays, a neutron spectrum similar to that encountered in Hiroshima at 1–1.5 km from the epicenter induced a severe metabolic dysregulation, with perturbations in amino acid metabolism and fatty acid β-oxidation being the predominant ones. Urinary metabolites were able to discriminate between neutron and X rays on day 1 as well as day 7 postirradiation, while serum markers showed such discrimination only on day 1. Free fatty acids from omega-6 and omega-3 pathways were also decreased with 1 Gy of neutrons, implicating cell membrane dysfunction and impaired phospholipid metabolism, which should otherwise lead to release of those molecules in circulation. While a precise relative biological effectiveness value could not be calculated from this study, the results are consistent with other published studies showing higher levels of damage from neutrons, demonstrated here by increased metabolic dysregulation. Metabolomics can therefore aid in identifying global perturbations in blood and urine, and effectively distinguishing between neutron and photon exposures.
INTRODUCTION
The increased threat of terrorist acts around the world has often been associated with increased probabilities of individuals or groups acquiring and using radioactive materials as improvised nuclear devices (IND) or radiological dispersal devices (RDD). Of particular concern is the detonation of an IND that would lead to high numbers of casualties and hundreds of thousands of survivors who will need to be evaluated qualitatively and quantitatively for exposure to radiation, including first responders and military personnel (1–3). Various biodosimetric methods have been proposed and evaluated for such scenarios (4, 5), with metabolomics gaining popularity in rapid assessment of biofluids. In particular, urine and serum have been very attractive and rich biofluids for such measurements, since their acquisition in large-scale events will be mostly noninvasive and can be achieved even from severely injured or unresponsive individuals.
Metabolomics involves the collective assessment of small molecules (<1 kDa) in a biofluid or tissue, providing a snapshot of the metabolism and allowing for development of a biosignature associated with specific stress- and injury-related exposures. A subcategory of metabolomics has emerged in the past decade, termed lipidomics, which is used to investigate collective changes in lipids such as free fatty acids and phospholipids, among others. As such, metabolomics and lipidomics have been utilized to create biosignatures in biofluids of rodent models, nonhuman primates and human populations, examining relative scenarios such as external and internal exposures, dose and dose-rate effects and different radiation qualities (6–19). An identified biomarker for biodosimetry refers to a measurable molecule in a given biofluid that indicates exposure to ionizing radiation and/or absorbed dose.
In the case of an IND, the immediate exposure will most likely involve a combination of photons with neutrons, while the majority of biological responses will be due to the neutrons, given the densely ionizing nature of those particles. This scenario has been encountered with the atomic bomb in Hiroshima, a focus of the Life Span Study undertaken by the Radiation Effects Research Foundation (RERF) to elucidate the long-term health risks associated with radiation exposure (20). Different relative biological effectiveness (RBE) values have been documented depending on whether a pure or mixed neutron field was used (20). RERF has used a constant RBE of 10, although evidence suggests that the RBE of a neutron dose decreases with increasing neutron and gamma-ray dose in a mixed field (20) and that a dose-dependent RBE may be more appropriate (21). However, a vast number of studies utilize a pure monoenergetic neutron source, an approach that does not adequately identify or mimic the radiation field observed in the atomic bomb incidents.
To simulate these particular conditions, an accelerator-based neutron irradiation facility was constructed at the Columbia University Radiological Research Accelerator Facility (RARAF; Irvington, NY) (22, 23). The spectrum of neutrons was similar to that encountered in Hiroshima at 1–1.5 km from the epicenter (24), and dosimetry measurements indicated that the neutron exposures contained a ~20% photon dose with neutron energies ranging between 0.2 and 9 MeV, described in detail by Xu, et al. (22). The doses were chosen based on the limited data on IND-spectrum neutrons (25, 26). A calculated RBE of 4, based on micronuclei formation for this particular mixed field (22), was used for the comparative doses in our current studies; namely the neutron doses used were 0.25 and 1 Gy, whereas the X-ray doses used were 1 and 4 Gy. Global metabolomic and lipidomic analyses were performed on urine and serum to identify biomarkers associated with different radiation qualities. Primary analysis was based on a comparison of 1 Gy neutrons with 1 Gy X rays as an initial attempt to identify any metabolic effects of different radiation qualities, and ultimately, to be able to identify the contribution of neutrons and/or photons during a mixed exposure with various levels of neutron composition. Neutron doses much higher than 1 Gy would not be expected outside the blast zone in an IND scenario (27), and are therefore not relevant for medical triage. Results indicate a pronounced dysregulation of metabolism after exposure to neutrons compared to X rays, with free fatty acids and amino acid metabolism intermediates exhibiting a striking decrease in excreted or circulating levels. Equidose analysis (1 Gy) showed a more pronounced altered phenotype with neutrons at day 1 in both urine and serum and to a lesser extent at day 7 for serum compared to day 1. To our knowledge, this is the first published study with a Hiroshima-like neutron spectrum to identify metabolic biomarkers specifically associated with different radiation qualities in easily accessible biofluids and to demonstrate the overall differences in the metabolome based on neutron vs. photon exposures.
MATERIALS AND METHODS
Chemicals
All chemicals used were of the highest purity, and reagents used were of LC-MS grade. All chemicals for the metabolomics studies (L-phenylalanine, phenylpyruvic acid, xanthurenic acid, decanoylcarnitine, sebacic acid, L-tysorine, L-glutamic acid, taurine, succinic acid, creatinine, L-carnitine, pyroglutamic acid, eicosapentaenoic acid, linoleic acid, arachidonic acid, uric acid and citric acid) were purchased from Sigma-Aldrich® (St. Louis, MO). Sphinganine-1-phosphate was purchased from Avanti® Polar Lipids, Inc. (Alabaster, AL), N-decanoyglycine from Hit2Lead (San Diego, CA) and decanoylcarnitine from Tocris Bioscience (Bristol, UK). Standards for lipidomics have been previously described elsewhere (9, 28) and covered all broad lipid classes.
Experimental Design and Sample Collection
All studies used C57Bl/6J male 8–9-week-old mice. At 7 weeks old, cage mates were purchased from Charles River Laboratories and kept in the animal facility for one week of adaptation before irradiation. Animals were housed three per cage, provided with food and water ad libitum and kept on a 12:12 h light:dark schedule. All experiments and experimental setups were approved by the Columbia University IACUC. Six mice were used for each group (10 groups total), except for the day 7, neutron 1 Gy irradiated group, consisting of 5 animals.
Urine was collected from the mice after they were placed in metabolic cages (cylindrical shape, size of the mouse chamber: 38 cm height, 17.5 cm diameter, mouse area = 240.4 cm2; Tecniplast®, Exton, PA) supplied with food and water. The urine was directed by funnel to small containers at the bottom of the metabolic cages, separating it from the fecal material. Before the experiments, mice were acclimated to the metabolic cage for one 24-h period. Mice were placed a second time in the metabolic cage for 24 h for preirradiation urine collection. Twenty-four hours later, the mice were irradiated and immediately placed back in the cages. Urine was collected at 24 h and 7 days postirradiation and frozen at −80°C until transfer to George-town University (Washington, DC).
Serum was collected through cardiac punctures. Blood (100 μl), was added to serum separator tubes (BD Microtainer® Tubes; Becton, Dickinson and Co., Franklin Lakes, NJ) and allowed to clot for a minimum of 30 min at room temperature. The tubes were then centrifuged at 12,000g for 5 min at 4°C, and the serum was transferred to Eppendorf tubes and stored immediately at −80°C until transfer to Georgetown University.
Irradiations and Dosimetry
Neutron irradiations were performed at RARAF, using an accelerator-based neutron irradiator mimicking the neutron energy spectrum from an IND (22). Briefly, a mixed beam of atomic and molecular ions of hydrogen and deuterium were accelerated to 5 MV potential and used to bombard a thick beryllium target. The energy spectrum of neutrons emitted at 60° to the ion-beam axis closely mimics the Hiroshima spectrum at 1–1.5 km from the epicenter (23). During irradiation, between 6 and 18 mice were placed in adjacent positions on an 18-position Ferris wheel, rotating around the beryllium target, at an angle of 60° to the particle beam and a distance of 17.5 cm from the beryllium target. Mice were placed in mouse holders, based on standard 50-ml conical centrifuge tubes. For training purposes, on days 2 and 4 before irradiation, the mice were placed in the irradiation holders for 10 and 30 min, respectively. These holders were used for both neutron and X-ray irradiations. The mouse holders are designed to maintain a constant horizontal orientation as the wheel rotates, providing an isotropic irradiation, while maintaining the mice in an upright orientation, reducing stress. The wheel is rotated at a speed of approximately 2 min per revolution and the dose rate adjusted so that the minimal dose is delivered in 10 rotations (20 min) with the mouse holders flipped end-to-end halfway through, so that the front and back of the mouse receive equivalent doses. To ensure a uniform scatter dose, when fewer than 18 mice were on the wheel, four 50 ml tubes containing Lucite phantoms were placed on the wheel, two at either end of the string of mouse holders.
Irradiations were performed with a total beam current of 18 μA, resulting in a dose rate of 1.55 Gy/h of neutrons and 0.4 Gy/h of gamma rays. Further details on dosimetry can be found elsewhere (22). X-ray irradiations were performed using a Westinghouse Coronado orthovoltage X-ray machine at 250 kVp and 15 mA and using a 0.5-mm copper plus 1-mm aluminum filter. Dose rate at the mouse location was 1.23 Gy/min, as determined using a Victoreen Model 570 Condenser R-Meter with a 250r chamber. No radiotoxicity was observed in the mice from any of the neutrons or X-ray doses with regards to survival until day 7.
Sample Preparation and Profiling
For metabolomic analysis, urine samples were deproteinated with 50:50 acetonitrile:water in a 1:5 dilution (20 μl of urine used) and serum samples were deproteinated with 66:34 acetonitrile:water in a 1:40 dilution (5 μl of serum used). Serum samples were incubated on ice for 10 min. Internal standards consisted of 4 μM debrisoquine sulfate and 30 μM 4-nitrobenzoic acid. Both serum and urine samples were centrifuged for 20 min (4°C maximum speed). For serum lipidomic analysis, samples were prepared with chloroform:methanol (2:1) with the appropriate lipidomic internal standards (28) (25 μl of serum used). Two microliters of each sample were injected in a Waters® Acquity Ultra Performance Liquid Chromatography (UPLC®) system, coupled to a Waters Xevo® G2 QTOF-MS (Waters, Milford MA). A BEH C18 column (130 Å, 1.7 μm, 2.1 × 50 mm, 40°C for urine and 60°C for serum) was used for the urine and serum metabolomics, while a CSH C18 column (130 Å, 1.7 μm, 2.1 × 100 mm) was used for the lipidomic analysis. All analyses were performed in both positive and negative electrospray ionization modes, with MSE function. The chromatographic and mass spectrometry conditions are provided in Supplementary Table S1 (http://dx.doi.org/10.1667/RR14656.1.S1). Quality control samples were also created from pooled samples and were run every 10 samples to assess for retention time drift and chromatographic quality.
Data Processing and Analysis
Peak alignment and deconvolution for the global urine analysis was performed with MarkerLynx™ XS software (Waters), with normalization applied so that the sum of the marker intensities for each sample would add to 10,000. To assess for differences in glomerular filtration rates, each sample was further normalized to its respective creatinine level ([M + H]+ = 114.0667, retention time 0.36 min). Serum metabolomic and lipidomic preprocessing was performed with Progenesis QI (Nonlinear Dynamics, Newcastle, UK). Normalization was performed with the option, “normalize to all compounds”, utilized in previously published studies (15, 28, 29) to minimize the influence of outliers and create a normal distribution of the data. Deconvoluted data consisted of a matrix with normalized abundance levels for each ion of each sample, where each ion was identified by a unique set of mass over charge ratios (m/z) and retention time.
Multivariate statistical analysis was conducted in serum and urine with SIMCA-P+ software version 13 (Umetrics, Umea, Sweden) to generate principal component analysis (PCA) score plots of the first two components [t1] and [t2]. Univariate statistical analysis was performed on the neutron vs. X ray groups (1 Gy) at days 1 and 7 postirradiation with the in-house statistical software, MetaboLyzer (30). For complete presence ions (≥75% presence in each group), statistically significant ions (P < 0.05) were identified with Welch’s t test with a false discovery rate (FDR) of 0.2, while statistically significant (P < 0.05) partial presence ions (<75% presence in each group) with FDR = 0.2 were identified with the categorical Barnard’s test. Putative identities of the ions were obtained through the databases HMDB (31), KEGG (32, 33) and LIPID MAPS® (34, 35) with mass accuracy cutoff of 10 (ppm error). Volcano plots (log-fold change vs. –log FDR corrected P value) were constructed from the complete presence ions to identify changes in individual ions, where grey dots (one dot represents a single ion) signify no change and red dots signify significant changes. Putative lipidomic identities were further assigned through databases such as LIPID MAPS and LipidBlast (36) directly incorporated into Progenesis QI, with a ppm error of 10.
Definitive Identification of Putative Metabolites and Statistical Analysis
Definitive identification of putative identities was performed with tandem mass spectrometry against pure chemicals. Ramping collision energy from 0 to 40 eV was used to fragment each putative m/z and pure chemical, with retention time utilized as an additional identifier. For lipidomics, utilization of fragmentation through the MSE function, together with specific elution times of the internal standards, allowed for lipid class assignment of the statistically significant ions through Progenesis QI. Definitive identities to individual lipids through pure chemicals were not obtained, therefore, data are presented in class plus carbon: double-bond content format. Examples of the raw chromatograms and MS/MS spectra are shown in Supplementary Fig. S1 (http://dx.doi.org/10.1667/RR14656.1.S1). All fragmentation patterns were cross referenced to those at the online database METLIN™ (Scripps, La Jolla, CA) (37).
All data were presented as mean ± standard error of the mean (SEM). Important markers were determined from 1 Gy comparisons and their levels further determined in other doses, including controls, and time points. Statistical differences between controls and all exposures for each time point were determined with one-way analysis of variance (ANOVA), with P < 0.05 considered significant. Statistical analysis between two groups (controls vs. 1 Gy neutrons, controls vs. 1 Gy X rays, 1 Gy neutrons vs. 1 Gy X rays) was performed with Welch’s t test, with P < 0.05 considered significant. Fold changes were calculated as experimental group divided by control group. When fold change (FC) was <1, it was substituted with −(1/FC). Fold changes <1.2 were depicted as a decreased trend (↓), while fold changes >1.2 were depicted as an increased trend (↑).
Receiver Operating Characteristic Curves and Heatmaps
Receiver operating characteristic (ROC) curves of the definitively identified metabolites as a signature was performed through MetaboAnalyst version 3.0 (McGill University, Montreal, Canada) (38). Features with >75% missing values were removed from the analysis. No normalization, transformation or scaling of the data was conducted. A multivariate ROC curve was generated with the multivariate algorithm random forests, utilizing a combination of biomarkers with Monte Carlo cross-validation and balanced subsampling. Briefly, two-thirds of the samples were randomly selected, a classification model was built on the identified important features, and the remaining one-third were used for validation of the model. An area under the curve (AUC) value of >0.9 represents an excellent classification model, and an AUC value of 0.8–0.9 represents a good classification model. Biomarkers were ranked by mean importance through random forests classification. Graphical representation of the average importance of each marker was also generated through MetaboAnalyst 3.0. Red and green represent high and low importance, respectively, between the 1 Gy comparisons. Heatmaps of the definitively identified metabolites with the normalized abundance were also generated through MetaboAnalyst 3.0. Data filtering was based on standard deviation. A Euclidean distance measurement was utilized, with Ward as the clustering algorithm. Green represents relative decreased levels, while red represents the opposite.
RESULTS
Mice were irradiated with either neutrons or X rays in the same facility. Animal weight was recorded, with no statistically significant differences observed among the different groups (Supplementary Fig. S2A; http://dx.doi.org/10.1667/RR14656.1.S1). No statistical significance was observed in creatinine levels among the groups in each time point (Supplementary Fig. S2B), thus abundance levels of each ion in each sample were normalized to the respective sample creatinine levels. Multivariate data analysis showed distinct metabolic differences through clustering of the groups, depicted with the generation of representative PCA score plots for ESI− in urine and ESI+ in serum (Fig. 1A, Supplementary Fig. S3; controls included for all analyses). In particular, metabolomic analysis of ESI− urine showed distinct clustering of each 1 Gy group in each time point, whereas in ESI+ serum only the day 1 samples formed unique clusters (Fig. 1A). No samples were found outside the Hotelling’s T2 tolerance ellipse. For urine, the goodness of fit, R2X, equaled 0.503 and goodness of prediction, Q2X, equaled 0.228, while for serum the values were increased to R2X = 0.666 and Q2X = 0.568. The values for all other analyses (including the controls) showed similar levels for the R2X and Q2X parameters (Supplementary Fig. S1A–F; http://dx.doi.org/10.1667/RR14656.1.S1). Evaluation of the patterns of statistically significant ions [as shown in the volcano plots for both time points (Fig. 1B)] with merged data from both ESI+ and ESI− showed a far greater response of individual ions at day 7 in urine, whereas serum exhibited a higher response at day 1. Regarding the lipidomic results, a limited number of ions were statistically significant at day 1, and none at day 7 after FDR correction (data not shown). Therefore, urine metabolomics are suitable in this case for determination of exposure to different radiation qualities in both time points, whereas serum is more informative at the earlier time point.
FIG. 1.
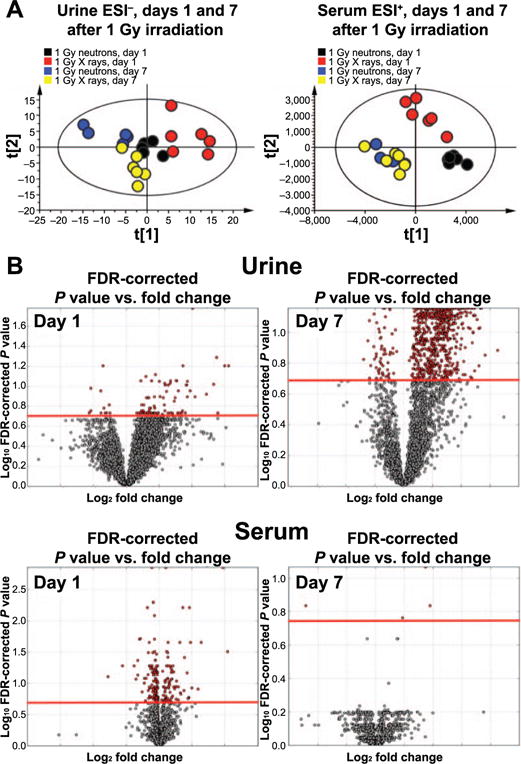
Multivariate and univariate data analysis of urine and serum metabolomics. Panel A: PCA analysis of urine ESI− data reveals distinct clustering of all 1 Gy groups, with time separation on component 1. Serum ESI+ data show time-dependent separation, with unique metabolic profiles for 1 Gy neutrons vs. X rays at day 1 only. Panel B: Volcano plot analysis for urine shows ions that are significantly perturbed on both days 1 and 7 postirradiation (red), while serum metabolomics is more informative at day 1.
Univariate data analysis of the urinary 1 Gy groups (neutrons vs. X rays) revealed 6 ions with P < 0.05 [Welch’s t test (FDR = 0.2)] that were definitively identified with tandem mass spectrometry against pure chemicals. These included L-phenylalanine, phenylpyruvic acid and N-decanoylglycine for day 1. Xanthurenic acid, phenylpyruvic acid, decanoylcarnitine and sebacic acid were identified for day 7 (Fig. 2). Table 1 shows the fold changes of 1 Gy irradiation compared to controls or 1 Gy comparisons with a generalized decreased trend (lower levels in neutron compared to X-ray radiation group) and proposed biochemical pathway involvement. Supplementary Table S2 shows the mean ± SEM values in each group and ANOVA P values of intergroup statistical analysis.
FIG. 2.
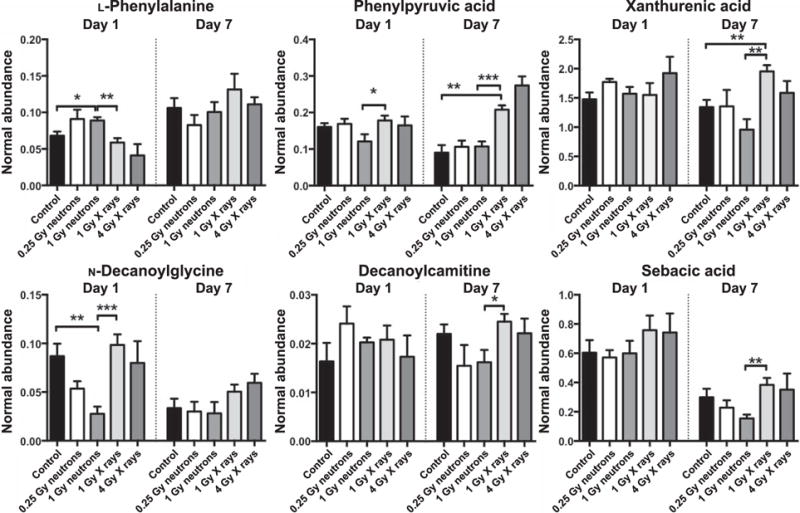
Urinary metabolic markers at days 1 and 7 postirradiation, based on comparison between equidoses and radiation type. L-phenylalanine, phenylpyruvic acid and N-decanoylglycine exhibited statistical significance when comparing 1 Gy neutrons vs. 1 Gy X rays at day 1. Phenylpyruvic acid, xanthurenic acid, decanoylcarnitine and sebacic acid were significantly different between the equidose groups at day 7. All data are presented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 (Welch’s t test).
TABLE 1.
Fold Changes and Trends of Urine Metabolomics Data
| Metabolite | m/z_RT | Proposed pathway/description | ESI and adduct | ppm error | 1 Gy neutron/control | 1 Gy X ray/control | 1 Gy neutron/1 Gy X ray | Trend between 1 Gy exposed (neutron/X ray) 1 | P value 1 Gy comparison | FDR value |
|---|---|---|---|---|---|---|---|---|---|---|
| Day 1 | ||||||||||
| L-phenylalanine | 164.0701_0.79 | Amino acid | [M − H]− | 9.7 | 1.3 | −1.2 | 1.5 | é | 0.003 | 0.18 |
| Phenylpyruvic acid | 163.0393_1.71 | Phenylalanine metabolism | [M − H]− | 4.7 | −1.3 | 1.1 | −1.5 | ê | 0.04 | 0.27 |
| Xanthurenic acid | 206.0454_1.26 | Tryptophan metabolism | [M + H]+ | 3 | 1.1 | 1.1 | 1.0 | – | 0.93 | 0.97 |
| N-decanoylglycine | 228.1592_6.00 | Fatty acid metabolism | [M + H]+ | 5.8 | −3.1 | 1.1 | −3.6 | ê | <0.001 | 0.10 |
| Decanoylcarnitine | 316.2464_6.12 | Fatty acid β-oxidation | [M + H]+ | 5.9 | 1.2 | 1.3 | 1.0 | – | 0.87 | 0.94 |
| Sebacic acid | 203.1286_5.34 | Dicarboxylic acid | [M + H]+ | 4 | 1.0 | 1.3 | −1.3 | ê | 0.26 | 0.52 |
| Day 7 | ||||||||||
| L-phenylalanine | 164.0701_0.79 | Amino acid | [M − H]− | 9.7 | −1.1 | 1.2 | −1.3 | ê | 0.25 | 0.54 |
| Phenylpyruvic acid | 163.0393_1.71 | Phenylalanine metabolism | [M − H]− | 4.7 | 1.2 | 2.3 | −1.9 | ê | <0.001 | 0.07 |
| Xanthurenic acid | 206.0454_1.26 | Tryptophan metabolism | [M + H]+ | 3 | −1.4 | 1.5 | −2.0 | ê | 0.002 | 0.07 |
| N-decanoylglycine | 228.1592_6.00 | Fatty acid metabolism | [M + H]+ | 5.8 | −1.2 | 1.5 | −1.8 | ê | 0.11 | 0.30 |
| Decanoylcarnitine | 316.2464_6.12 | Fatty acid β-oxidation | [M + H]+ | 5.9 | −1.4 | 1.1 | −1.5 | ê | 0.03 | 0.15 |
| Sebacic acid | 203.1286_5.34 | Dicarboxylic acid | [M + H]+ | 4 | −1.9 | 1.3 | −2.5 | ê | 0.003 | 0.07 |
Table 2 and Supplementary Table S3 show similar information for the serum metabolomics, with 12 metabolites definitively identified: L-tyrosine, L-glutamic acid, taurine, succinic acid, L-carnitine, pyroglutamic acid, linoleic acid, arachidonic acid, uric acid, sphinganine-1-phosphate, citric acid and eicosapentaenoic acid. Trends of the 1 Gy comparisons showed a generalized decrease at day 1 for neutrons compared to X rays, while day 7 had an approximately equal distribution of increased and decreased levels of metabolites in 1 Gy neutrons vs. 1 Gy X rays. L-Tyrosine, taurine, L-carnitine, uric acid, pyroglutamic acid and succinic acid exhibited statistically significant differences between the 1 Gy groups at day 1. Sphinganine-1-phosphate was the only metabolite that exhibited statistical significance between the 1 Gy groups at both time points, while no other metabolites showed statistical significance for 1 Gy irradiation at day 7. On the other hand, L-glutamic acid and citric acid were statistically significantly different only between controls and the 1 Gy neutron groups at day 1. L-carnitine was the only metabolite increased at day 1 in the 1 Gy neutron group compared to all groups, further implicating mitochondrial and energy metabolism in radiation responses. These results are shown in Table 2 and Supplementary Fig. S4 (http://dx.doi.org/10.1667/RR14656.1.S1).
TABLE 2.
Fold Changes and Trends of Serum Metabolomics Data
| Metabolite | m/z_RT | ESI and adduct | Proposed pathway/description | ppm error | 1 Gy neutron/Control | 1 Gy X ray/Control | 1 Gy neutron/1 Gy X ray | Trend between 1 Gy exposed (neutron/X ray) | P value 1 Gy comparisons | FDR value |
|---|---|---|---|---|---|---|---|---|---|---|
| Day 1 | ||||||||||
| L-tyrosine | 182.0816_0.36 | [M + H]+ | Amino acid | 2.7 | −1.8 | −1.2 | −1.5 | ê | 0.04 | 0.23 |
| L-glutamic acid | 146.0460_0.34 | [M − H]− | Arginine biosynthesis and glutathione metabolism | 0.9 | −2.3 | −1.5 | −1.5 | ê | 0.09 | 0.34 |
| Taurine | 124.0068_0.33 | [M − H]− | Bile acid metabolism | 4.3 | −1.2 | 1.0 | −1.1 | – | 0.05 | 0.25 |
| Succinic acid | 117.0187_0.38 | [M − H]− | Dicarboxylic acid | 4.7 | −1.4 | 1.1 | −1.5 | ê | 0.01 | 0.12 |
| L-carnitine | 162.1129_0.33 | [M + H]+ | Fatty acid β-oxidation | 2.7 | 1.3 | 1.0 | 1.3 | é | 0.006 | 0.09 |
| Pyroglutamic acid | 128.0346_0.36 | [M − H]− | Glutathione metabolism | 4.8 | −2.0 | −1.3 | −1.6 | ê | 0.03 | 0.18 |
| Eisosapentaenoic acid | 303.2327_8.09 | [M + H]+ | Omega-3 fatty acid | 2.9 | −3.4 | 1.1 | −3.7 | ê | <0.001 | 0.02 |
| Linoleic acid | 279.2323_8.41 | [M − H]− | Omega-6 fatty acid | 2 | −4.8 | −1.5 | −3.2 | ê | 0.003 | 0.07 |
| Arachidonic acid | 303.2322_8.34 | [M − H]− | Polyunsaturated fatty acid | 2.4 | −1.5 | −1.2 | −1.3 | ê | 0.14 | 0.41 |
| Arachidonic acid | 305.2489_8.34 | [M + H]+ | Polyunsaturated fatty acid | 4.8 | −1.4 | −1.1 | −1.3 | ê | 0.02 | 0.17 |
| Uric acid | 167.0204_0.34 | [M − H]− | Pyrimidine metabolism | 3.5 | −1.4 | −1.1 | −1.4 | ê | 0.002 | 0.05 |
| Sphinganine-1-phosphate | 380.2566_7.10 | [M − H]− | Sphingolipid metabolism | 1.2 | 1.0 | −1.4 | 1.4 | é | 0.04 | 0.23 |
| Citric acid | 191.0192_0.38 | [M − H]− | TCA cycle | 2.7 | −1.5 | −1.3 | −1.2 | ê | 0.22 | 0.51 |
| Day 7 | ||||||||||
| L-tyrosine | 182.0816_0.36 | [M + H]+ | Amino acid | 2.7 | −1.1 | 1.0 | −1.2 | ê | 0.54 | 0.91 |
| L-glutamic acid | 146.0460_0.34 | [M − H]− | Arginine biosynthesis and glutathione metabolism | 0.9 | −1.6 | −1.1 | −1.4 | ê | 0.25 | 0.80 |
| Taurine | 124.0068_0.33 | [M − H]− | Bile acid metabolism | 4.3 | −1.6 | −1.2 | −1.3 | ê | 0.05 | 0.64 |
| Succinic acid | 117.0187_0.38 | [M − H]− | Dicarboxylic acid | 4.7 | −1.1 | −1.2 | 1.0 | – | 0.87 | 0.99 |
| L-carnitine | 162.1129_0.33 | [M + H]+ | Fatty acid β-oxidation | 2.7 | 1.1 | −1.1 | 1.1 | – | 0.05 | 0.65 |
| Pyroglutamic acid | 128.0346_0.36 | [M − H]− | Glutathione metabolism | 4.8 | −1.2 | −1.4 | 1.1 | – | 0.49 | 0.90 |
| Eisosapentaenoic acid | 303.2327_8.09 | [M + H]+ | Omega-3 fatty acid | 2.9 | 1.3 | −1.6 | 2.1 | é | 0.03 | 0.64 |
| Linoleic acid | 279.2323_8.41 | [M − H]− | Omega-6 fatty acid | 2 | 1.0 | −1.4 | 1.4 | é | 0.52 | 0.90 |
| Arachidonic acid | 303.2322_8.34 | [M − H]− | Polyunsaturated fatty acid | 2.4 | 1.3 | −1.1 | 1.4 | é | 0.18 | 0.76 |
| Arachidonic acid | 305.2489_8.34 | [M + H]+ | Polyunsaturated fatty acid | 4.8 | 1.2 | −1.1 | 1.3 | é | 0.09 | 0.69 |
| Uric acid | 167.0204_0.34 | [M − H]− | Pyrimidine metabolism | 3.5 | −1.3 | −1.2 | −1.1 | – | 0.45 | 0.90 |
| Sphinganine-1-phosphate | 380.2566_7.10 | [M − H]− | Sphingolipid metabolism | 1.2 | −1.5 | 1.1 | −1.6 | ê | 0.05 | 0.64 |
| Citric acid | 191.0192_0.38 | [M − H]− | TCA cycle | 2.7 | −1.1 | 1.0 | −1.1 | – | 0.43 | 0.76 |
Lipidomic analysis results are shown in Table 3 and Supplementary Table S4. Lipid classes and carbon:-double-bond content were assigned according to searches through databases with an additional level of verification through retention time aided by incorporation of internal standards, overall leading to 27 lipids found to be statistically significant. LysoPCs were further identified through fragmentation patterns of MSE with the primary product ion of m/z 184. All LysoPEs and free fatty acids through the lipidomic analysis remained putative, however, the retention time with the elution of pure standards narrowed down their identities and class. All cholesteryl esters and triacylglycerides showed significantly decreased levels at day 1 in the 1 Gy neutron group compared to 1 Gy X-ray group, while levels of cholesteryl esters also remained decreased at day 7, and triacylglycerides (TG) showed either no appreciable changes or were increased; however, fold changes did not exceed 1.3 with the exception of TG(55:3). The identified LysoPEs, LysoPCs and free fatty acids exhibited increased circulating levels in the 1 Gy neutron group vs. 1 Gy X-ray group at day 7, compared to day 1, when their levels were lower in the 1 Gy neutron group vs. 1 Gy X-ray group.
TABLE 3.
Fold Changes and Trends of Serum Lipidomics Data
| Metabolite | m/z_RT | ESI and adduct | ppm error | 1 Gy neutron/Control | 1 GyX ray/Control | 1 Gy neutron/1 Gy X ray | Trend between 1 Gy exposed (neutron/X ray) | P value 1 Gy comparisons | FDR value |
|---|---|---|---|---|---|---|---|---|---|
| Day 1 | |||||||||
| Linoleic acid | 279.2330_2.47 | [M − H]− | 0.5 | −3.2 | −1.4 | −2.2 | ê | <0.001 | 0.06 |
| Docosahexanoic acid | 327.2327_2.23 | [M − H]− | 0.6 | −1.8 | −1.2 | −1.5 | ê | <0.001 | 0.08 |
| Arachidonic acid | 303.2327_2.35 | [M − H]− | 0.8 | −1.8 | −1.2 | −1.5 | ê | 0.002 | 0.11 |
| LysoPC(18:0) | 524.3714_1.99 | [M + H]+ | 0.7 | −1.3 | 1.0 | −1.4 | ê | 0.002 | 0.12 |
| LysoPC(20:1) | 550.3873_2.08 | [M + H]+ | 1.2 | −3.0 | 1.0 | −2.9 | ê | <0.001 | 0.06 |
| LysoPE(20:0) | 510.3560_1.65 | [M + H]+ | 1.2 | −1.4 | 1.0 | −1.4 | ê | <0.001 | 0.06 |
| LysoPE(22:0) | 538.3871_2.367 | [M + H]+ | 0.7 | −1.4 | 1.1 | −1.6 | ê | 0.001 | 0.10 |
| CE(18:2) | 666.6187_7.93 | [M + NH4]+ | 0.6 | −1.2 | 1.0 | −1.3 | ê | <0.001 | 0.08 |
| CE(18:3) | 664.6029_7.77 | [M + NH4]+ | 0.3 | −1.5 | 1.1 | −1.6 | ê | 0.002 | 0.11 |
| CE(20:4) | 690.6185_7.82 | [M + NH4]+ | 0.4 | −1.2 | 1.1 | −1.3 | ê | <0.001 | 0.08 |
| CE(20:5) | 688.6029_7.66 | [M + NH4]+ | 0.3 | −1.7 | 1.1 | −1.8 | ê | 0.002 | 0.11 |
| CE(22:6) | 714.6186_7.74 | [M + NH4]+ | 0.4 | −1.3 | 1.1 | −1.4 | ê | 0.005 | 0.18 |
| TG(52:1) | 878.8167_8.15 | [M + NH4]+ | 0.4 | −3.3 | −1.9 | −1.8 | ê | 0.004 | 0.17 |
| TG(52:3) | 874.7860_7.85 | [M + NH4]+ | 0.3 | −1.9 | −1.1 | −1.8 | ê | 0.004 | 0.17 |
| TG(55:3) | 916.8323_7.66 | [M + NH4]+ | 0.5 | −3.1 | −1.2 | −2.7 | ê | 0.004 | 0.17 |
| TG(56:4) | 928.8320_8.02 | [M + NH4]+ | 0.8 | −3.0 | 1.3 | −4.0 | ê | 0.004 | 0.17 |
| TG(56:6) | 924.7988_7.80 | [M + NH4]+ | 2.9 | −2.9 | 1.0 | −2.9 | ê | <0.001 | 0.09 |
| TG(56:8) | 920.7704_7.56 | [M + NH4]+ | 0.3 | −2.6 | −1.1 | −2.5 | ê | <0.001 | 0.09 |
| TG(57:3) | 944.8642_7.85 | [M + NH4]+ | 0.2 | −1.7 | −1.1 | −1.6 | ê | 0.002 | 0.11 |
| TG(57:3) | 944.8642_8.21 | [M + NH4]+ | 0.3 | −1.8 | 1.0 | −1.8 | ê | 0.004 | 0.17 |
| TG(57:4) | 942.8486_7.69 | [M + NH4]+ | 0.4 | −1.6 | 1.0 | −1.6 | ê | 0.004 | 0.17 |
| TG(58:10) | 944.7705_7.43 | [M + NH4]+ | 0.5 | −3.8 | −1.1 | −3.5 | ê | 0.003 | 0.17 |
| TG(58:10) | 927.7430_7.72 | [M + H]+ | 0.7 | −8.1 | 1.0 | −8.2 | ê | 0.004 | 0.17 |
| TG(58:3) | 958.8798_7.85 | [M + NH4]+ | 0.1 | −1.9 | −1.1 | −1.8 | ê | 0.003 | 0.16 |
| TG(58:4) | 956.8632_7.69 | [M + NH4]+ | 0.8 | −1.9 | −1.1 | −1.8 | ê | 0.005 | 0.17 |
| TG(58:8) | 948.8019_7.74 | [M + NH4]+ | 0.6 | −3.0 | −1.1 | −2.8 | ê | <0.001 | 0.08 |
| TG(58:9) | 946.7854_7.57 | [M + NH4]+ | 0.4 | −3.7 | −1.1 | −3.3 | ê | 0.002 | 0.12 |
| TG(59:3) | 972.8954_8.00 | [M + NH4]+ | 0.1 | −2.1 | −1.2 | −1.7 | ê | 0.004 | 0.17 |
| TG(61:8) | 990.8487_7.57 | [M + NH4]+ | 0.4 | –5.2 | 1.0 | –5.4 | ê | 0.001 | 0.10 |
| Day 7 | |||||||||
| Linoleic acid | 279.2330_2.47 | [M − H]− | 0.5 | −1.3 | −2.2 | 1.6 | é | 0.37 | 0.77 |
| Docosahexanoic acid | 327.2327_2.23 | [M − H]− | 0.6 | −1.1 | −1.4 | 1.3 | é | 0.28 | 0.72 |
| Arachidonic acid | 303.2327_2.35 | [M − H]− | 0.8 | −1.1 | −1.3 | 1.2 | é | 0.38 | 0.78 |
| LysoPC(18:0) | 524.3714_1.99 | [M + H]+ | 0.7 | −1.1 | −1.4 | 1.3 | é | 0.05 | 0.48 |
| LysoPC(20:1) | 550.3873_2.08 | [M + H]+ | 1.2 | 1.0 | −1.3 | 1.3 | é | 0.28 | 0.72 |
| LysoPE(20:0) | 510.3560_1.65 | [M + H]+ | 1.2 | 1.0 | −1.3 | 1.2 | é | 0.17 | 0.62 |
| LysoPE(22:0) | 538.3871_2.367 | [M + H]+ | 0.7 | −1.3 | −1.5 | 1.2 | é | 0.29 | 0.72 |
| CE(18:2) | 666.6187_7.93 | [M + NH4]+ | 0.6 | −1.2 | 1.1 | −1.3 | ê | 0.11 | 0.55 |
| CE(18:3) | 664.6029_7.77 | [M + NH4]+ | 0.3 | −1.2 | 1.2 | −1.4 | ê | 0.03 | 0.48 |
| CE(20:4) | 690.6185_7.82 | [M + NH4]+ | 0.4 | −1.1 | 1.2 | −1.3 | ê | 0.03 | 0.48 |
| CE(20:5) | 688.6029_7.66 | [M + NH4]+ | 0.3 | 1.1 | 1.2 | −1.1 | – | 0.53 | 0.86 |
| CE(22:6) | 714.6186_7.74 | [M + NH4]+ | 0.4 | −1.3 | 1.2 | −1.5 | ê | 0.03 | 0.48 |
| TG(52:1) | 878.8167_8.15 | [M + NH4]+ | 0.4 | 1.4 | 1.6 | −1.1 | – | 0.53 | 0.86 |
| TG(52:3) | 874.7860_7.85 | [M + NH4]+ | 0.3 | 1.8 | 1.3 | 1.3 | é | 0.11 | 0.54 |
| TG(55:3) | 916.8323_7.66 | [M + NH4]+ | 0.5 | 2.2 | 1.3 | 1.7 | é | 0.03 | 0.48 |
| TG(56:4) | 928.8320_8.02 | [M + NH4]+ | 0.8 | 2.1 | 2.1 | 1.0 | – | 0.93 | 0.99 |
| TG(56:6) | 924.7988_7.80 | [M + NH4]+ | 2.9 | 1.9 | 1.5 | 1.3 | é | 0.14 | 0.59 |
| TG(56:8) | 920.7704_7.56 | [M + NH4]+ | 0.3 | 1.5 | 1.3 | 1.2 | é | 0.26 | 0.71 |
| TG(57:3) | 944.8642_7.85 | [M + NH4]+ | 0.2 | 1.7 | 1.3 | 1.3 | é | 0.07 | 0.50 |
| TG(57:3) | 944.8642_8.21 | [M + NH4]+ | 0.3 | −0.6 | 1.7 | −0.7 | – | 0.003 | 0.48 |
| TG(57:4) | 942.8486_7.69 | [M + NH4]+ | 0.4 | 1.4 | 1.1 | 1.3 | é | 0.04 | 0.48 |
| TG(58:10) | 944.7705_7.43 | [M + NH4]+ | 0.5 | 1.5 | 1.4 | 1.1 | – | 0.39 | 0.78 |
| TG(58:10) | 927.7430_7.72 | [M + H]+ | 0.7 | 2.2 | 1.8 | 1.3 | é | 0.25 | 0.71 |
| TG(58:3) | 958.8798_7.85 | [M + NH4]+ | 0.1 | 1.8 | 1.3 | 1.3 | é | 0.10 | 0.53 |
| TG(58:4) | 956.8632_7.69 | [M + NH4]+ | 0.8 | 1.6 | 1.2 | 1.3 | é | 0.08 | 0.51 |
| TG(58:8) | 948.8019_7.74 | [M + NH4]+ | 0.6 | 2.0 | 1.8 | 1.1 | – | 0.52 | 0.86 |
| TG(58:9) | 946.7854_7.57 | [M + NH4]+ | 0.4 | 1.7 | 1.5 | 1.1 | – | 0.40 | 0.79 |
| TG(59:3) | 972.8954_8.00 | [M + NH4]+ | 0.1 | 1.7 | 1.7 | 1.0 | – | 0.99 | 1.00 |
| TG(61:8) | 990.8487_7.57 | [M + NH4]+ | 0.4 | 1.6 | 1.3 | 1.2 | é | 0.21 | 0.66 |
In terms of inflammatory mediators, identification of four omega-6 and omega-3 free fatty acids (linoleic acid, arachidonic acid, eisosapentaenoic acid and putative docosahexanoic acid) (Fig. 3), showed decreased levels of all metabolites at day 1 after 1 Gy neutron irradiation when compared to the controls or 1 Gy X-ray irradiation. Limited changes, however, were observed at day 7, with both eicosapentaenoic acid and putative docosahexanoic acid exhibiting decreased levels in the 1 Gy X-ray group compared to the controls and/or 1 Gy neutron group. The intermediate dihomo-γ-linolenic acid (DGLA) of the omega-6-related pathway and precursor α-linolenic acid were not identified in our analysis. Taken together, combining markers that exhibit significant differences between two radiation qualities could aid in the potential construction of a biosignature that would provide information on the radiation quality of the exposed person.
FIG. 3.
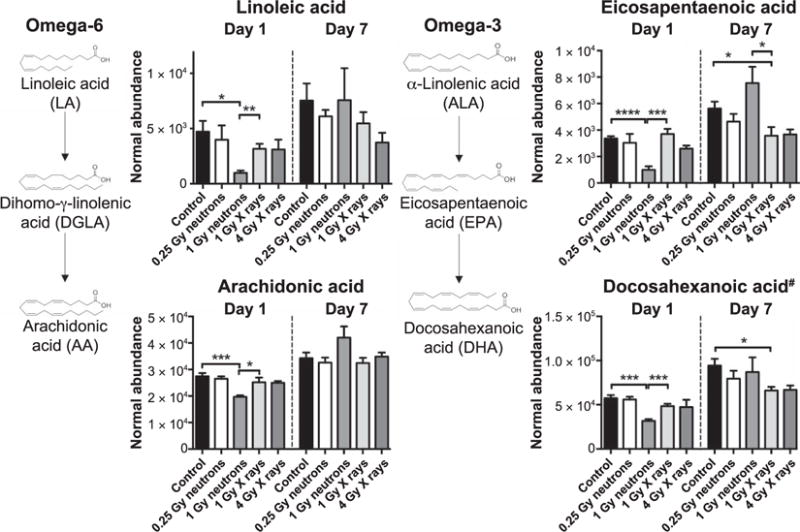
Alterations in free fatty acids of the omega-6 and omega-3 pathways (linoleic acid, arachidonic acid, eicosapentaenoic acid and putative docosahexanoic acid, designated with the symbol #). Neutron exposure (1 Gy) translates to general suppression of levels at day 1. Levels return to no statistical significance at day 7 compared to 1 Gy X-ray exposure. Eicosapentaenoic acid, an omega-3 fatty acid, is the only free fatty acid that reverses its circulating profile (increased compared to 1 Gy X rays) at day 7. All data are presented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 (Welch’s t test).
Based on the identified signatures in urine (metabolomics) and serum (metabolomics combined with lipidomics), ROC curves were constructed for the two time points by comparing the two 1 Gy groups to identify the discriminatory nature of the signature (sensitivity vs. specificity). As shown in Fig. 4 for urine, combinations of identified biomarkers from 2 to 6 provided excellent discriminatory power for both days with AUCs ranging from 0.972 to 1. The average importance of the metabolites is shown in Supplementary Fig. S5A (http://dx.doi.org/10.1667/RR14656.1.S1). In serum, however, the model utilizing up to 21 biomarkers (triacylglycerides and cholesteryl esters were excluded from the signature) exhibited high specificity and sensitivity only at day 1 (AUCs of 0.98–1), whereas at day 7 the identified metabolites failed to provide adequate discrimination between the two groups (AUCs of 0.708–0.8). The average importance of the metabolites is shown in Supplementary Fig. S4A. The patterns of change of the combined signatures (urine and serum) are further represented in a heatmap (Supplementary Fig. S5B), where at day 1, metabolite levels for the 1 Gy neutron group clearly showed a decrease compared to the 1 Gy X-ray group; however, at day 7 a roughly equal distribution of increased and decreased levels existed between the two groups. The AUC values for each individual metabolite based on individually constructed ROC curves are shown in Supplementary Table S5.
FIG. 4.
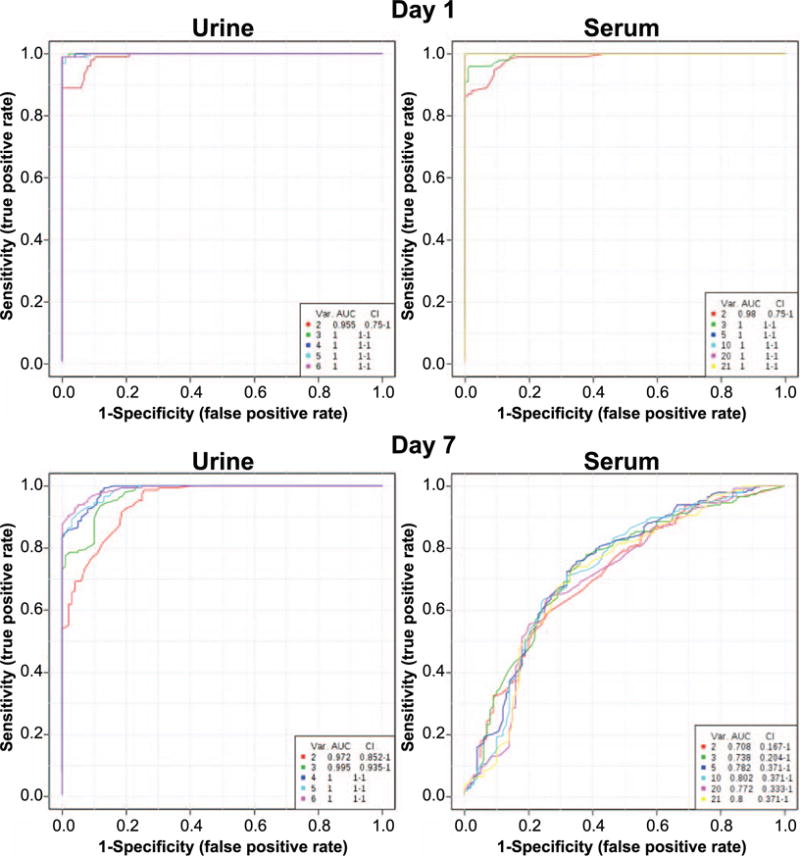
ROC curves based on definitively identified metabolites and lipidomics results. AUC values of >0.9 indicate a high specificity and sensitivity of the combination of metabolites for selective identification based on different radiation quality. The six metabolites identified in urine (L-phenylalanine, N-decanoylglycine, phenylpyruvic acid, sebacic acid, xanthurenic acid and decanoylcarnitine) provide a separation of 1 Gy neutron from 1 Gy X rays with high specificity and sensitivity at both days 1 and 7 postirradiation, as demonstrated by the high arch of the curve. The serum signature, on the other hand (metabolomics and lipidomics markers), exhibited high specificity and sensitivity only on day 1, with generally low AUC levels at day 7.
To calculate the overall severity of radiation type on the metabolome we utilized the results from MetaboLyzer, by combining the both presence (Welch’s t test results) and one presence (Barnard’s test) statistically significant ions (Fig. 5). Ratios revealed the effect of radiation type on the urinary metabolome. As shown in Fig. 5A, neutron exposure compared to controls showed a 4.1–5.2 times higher level of metabolic dysregulation compared to X-ray exposure at day 1 in all biofluids analyzed. Similar patterns persisted at day 7, although the levels of dysregulation were lower (1.7 ratio) for urinary metabolomics and lipidomic analysis. Serum metabolomic analysis, on the other hand, exhibited a similar response between the neutron and X-ray equidose groups (0.9 ratio). Determination of significant ions in each radiation vs. control group, and subsequent comparison of the assumed equi-responsive doses of different radiation quality, implied that the assumed RBE of 4 might not be adequately descriptive of the overall metabolomic responses. Comparing the 0.25 Gy neutron group to 1 Gy X-ray group, and 1 Gy neutron group to 4 Gy X-ray group (Fig. 5B), the values representative for the biological changes range between 1.5 to 7.1 for day 1 and 0.4 to 1.3 for day 7.
FIG. 5.
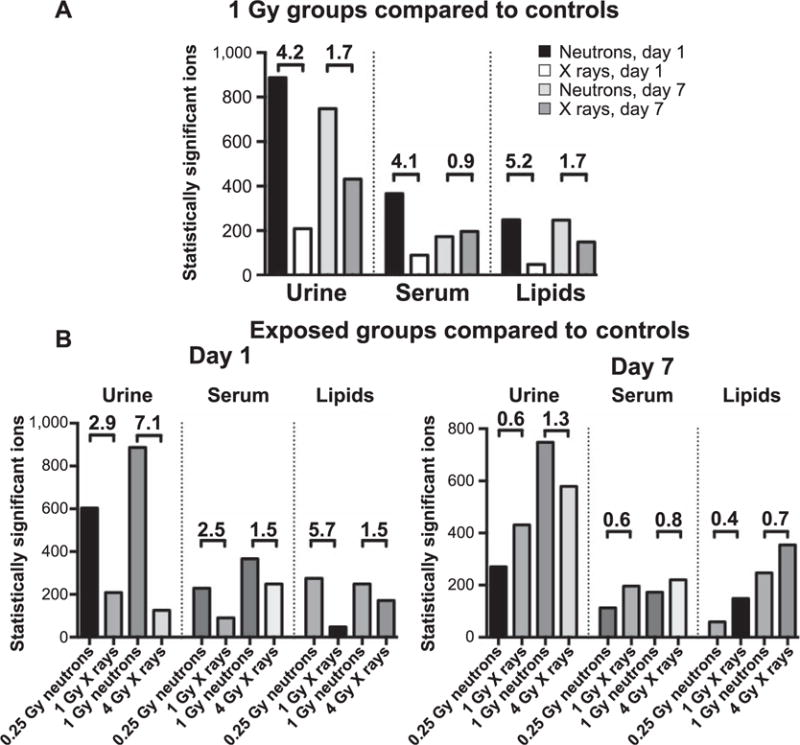
Comparison of the total number of statistically significant ions (Welch’s t test and Barnard’s test) among different biofluids, doses and time points. All differences in both panels were calculated as N/X. Panel A: Equidose analysis between neutrons and X rays. Neutron irradiation led to severe metabolic dysregulation, more pronounced in urine (4.2-fold difference at day 1 and 1.7 at day 7). Serum metabolomics showed similar patterns with high fold differences at day 1 (4.1 for metabolomics and 5.2 for lipidomics), decreasing levels to 0.9–1.7 at day 7. Panel B: The number of statistically significant ions was determined through univariate analysis of dose to controls and combining the number identified through Welch’s test and Barnard’s test. Fold changes as high as 7.1 at day 1 and increased metabolic perturbations in the neutron irradiated groups further show that neutrons have a higher biological effect compared to photons. Such dramatic effects disappeared by day 7, with fold changes ranging from 0.4 to 1.3
DISCUSSION
While a large number of casualties will occur either immediately or as a result of other injuries (e.g., trauma, burns) after detonation of an IND, a larger population will need to be evaluated rapidly for radiation exposure to determine whether medical intervention is required. In the case of an IND, neutrons will account for a portion of the radiation dose and significantly contribute to the biological effect, as has previously been determined in the atomic bombs in Hiroshima and Nagasaki. Therefore, it will be essential to discriminate between radiation types and subsequent biological effects. At RARAF, we constructed an accelerator-based neutron source mimicking the Hiroshima spectrum (22) and utilized metabolomics for assessing urine and serum of mice to identify metabolic differences between radiation types as a means of constructing potential biosignatures. The analysis was based on differences among 1 Gy exposed groups, to investigate whether the metabolic responses may differ based on radiation quality. In fact, a panel of metabolites for urine showed increased sensitivity and specificity for both time points, while serum markers were informative only at day 1 postirradiation (Fig. 4).
From the analysis obtained, it was evident that the overall potential metabolic pathways affected exhibited similarities with previously published analyses (9, 13, 29) between the two radiation types, with amino acids and fatty acid β-oxidation among the more prominent ones. Although DNA damage and repair products have been identified as significantly increased with neutrons, whether that is assessed with micronuclei (22), comet assay (39) or chromosomal changes (40, 41), small molecules associated with such a process or DNA repair were not identified in the biofluids through a global LC-MS approach. The exception was uric acid, which was previously identified in higher levels in urine of exposed mice (42) and humans (12). With 1 Gy exposure, no changes were observed in the X-ray group compared to the controls in the definitively identified metabolites, however, a significant decrease was observed in the 1 Gy neutron group. In fact, the overall patterns of metabolic changes were skewed towards more severe decreased levels in the neutron group, for both urine and serum at day 1 postirradiation when comparing the equidoses. This pattern was persistent in urine at day 7 postirradiation, whereas serum patterns showed an equal distribution of increased and decreased levels of metabolites. This collective analysis reiterates the already partially characterized biological significance of neutrons, that as densely ionizing radiation they are more damaging to a cell. This becomes more evident when comparing the total statistically significant ions between the two different radiation types, with regards to controls (Fig. 5A). A 4.1-to 5.2-fold change demonstrates the effectiveness of neutrons in delivering damage to cells and tissues, as reflected by dysregulation of metabolism, with attenuation of those effects in the later time point.
However, it should be noted that the particular biological effects of a pure neutron field on metabolism remains unknown, since the current beam contained ~20% of gamma rays. It is expected that pure neutrons may have a more severe effect on the metabolome, however, in this case a synergistic effect cannot be ruled out as leading to a more severe phenotype. Although an RBE cannot be calculated for metabolic responses due to the limited number of doses, other published work by Xu et al., with the same neutron field and quantifying DNA damage through mitotic micronuclei formation, demonstrated an RBE of 4 for neutrons (22). However, limited comparison of doses with an assumed RBE of 4 (Fig. 5B) suggests that the actual RBE values for global metabolic responses may be much higher, although in fact the RBE values for individual metabolites may differ from one to another.
Lipidomic analysis and individual free fatty acid identification through metabolomics of omega-6 and omega-3 precursors showed a significant decrease at day 1 in the high-dose neutron irradiated samples compared to controls or X-ray irradiated samples. No significant differences were observed in controls vs. X-ray group, which is in agreement with the published literature on serum from gamma-irradiated mice (13). Although a distinct possibility that reduced food intake may be responsible for the reduced levels, also seen through decreased triacylglyceride levels, the weight of the mice was not altered at day 1 or 7. In fact, at day 7 postirradiation, the levels of these lipids either remained stable between the groups or were increased. Early decrease of fatty acids, such as arachidonic acid, can be attributed directly to increased oxidative stress, which can lead to lipid peroxidation and protein alterations (43, 44). In addition, others have shown that membrane fluidity is decreased even with low doses of neutrons (0.9 cGy) and decreased lipid-to-protein ratios in membranes may be a consequence of increased lipid peroxidation and degradation (39). Changes in membrane solubilization and osmotic fragility after exposure to fast neutrons were also observed in erythrocytes (45). Similar results have been observed in gamma radiation exposures, with increased lipid peroxidation and alterations in the lipid content of the cell membrane after increased oxidative stress (46). Therefore, a change of phospholipids that can be converted to arachidonic acid through phospholipase A2 may be severely affected in the high-dose neutron group of this study, with downstream effects of oxylipin generation and immune system regulation. This was demonstrated in a blood gene expression-profiling study that identified enrichment in processes involved in lipid biosynthesis and metabolism in 1 Gy X-ray group vs. 1 Gy neutron group at day 1 but not day 7 postirradiation (25). On the other hand, increase of those markers at the later time point, whether statistically significant or suggestive of increased levels, indicate the existence of a delicate balance between a pro- and anti-inflammatory state implicating the immune system, as has been previously discussed (13, 47, 48). However, it remains to be determined what the downstream products of the omega-6 and omega-3 pathway are after exposure to neutrons, as the phenotype after gamma exposure clearly indicated the existence of a pro-inflammatory mechanism (13). Overall, phospholipid dysregulation has been proposed as a method to assess levels of external exposure (13, 49), and could be further explored in terms of radiation quality. As an exception to the overall suppression of circulating lipids after 1 Gy neutron irradiation, sphinganine-1-phosphate exhibits reduced circulating levels in the serum of the lower doses (0.25 Gy neutrons and 1 Gy X ray). The predominant difference between the equidoses is evident at day 7. Reduced circulating levels of this molecule, being an important intermediate in glycosphingolipid and sphingolipid metabolism, additionally implicates cell membrane regulation as dysregulated by neutrons. Additional analysis of lipid content of plasma membranes from tissues or circulating cells will provide important answers regarding membrane fluidity and levels of lipid peroxidation.
Finally, it is important to start recognizing that different patterns of circulating markers can indicate not only the dose that a person has been exposed to, but also the contribution of different radiation qualities, and whether an internal emitter, such as cesium-137 (137Cs) or strontium-90, may further contribute to the biological effects. In an IND scenario, individuals can be exposed either to external radiation and/or through ingestion/inhalation of radioactive materials that can contribute significantly to their overall dose. Medical treatment of those individuals will therefore greatly depend on correct assessment of their type of exposure. L-carnitine provides such an example that can be utilized to determine the specific radiation scenario. In our study, circulating free carnitine was increased at day 1 specifically after exposure to 1 Gy of neutrons, while levels remained unaltered with all other irradiated groups compared to the control group. Levels returned to normal by day 7. This is in direct contrast to serum-free carnitine levels after internal 137Cs exposure, which showed a persistent decrease for at least 30 days postirradiation, with accumulated doses varying from 1.95 Gy at day 2 to 9.91 Gy at day 30 (9). In a nonhuman primate model, free carnitine at day 7 after total-body gamma irradiation showed persistent increases in high doses associated with hematopoietic and/or gastrointestinal syndromes, while levels remained unaltered at the lower doses (50). While not different from the results obtained in this study for this particular metabolite at day 7, presence or absence of other markers in the samples, such as phenylpyruvic acid or uric acid, may provide the signature signal to identify the radiation quality and therefore lead to more detailed dose reconstruction. It is therefore possible to construct a panel of metabolites from the various radiation exposure scenarios, and based on the levels of select metabolites, guide the emergency personnel towards the appropriate treatment for each individual, such as chelators (e.g., Prussian blue), cytokine therapy or palliative care. Therefore, metabolomics provides a unique method to assess biodose and distinguish among radiation exposures such as different radiation types or internal vs. external exposures.
Supplementary Material
Fig. S1. Examples of raw chromatograms for lipidomics, serum metabolomics and urine metabolomics.
Fig. S2. Weight and creatinine levels of control and irradiated mice from each group.
Fig. S3. Expanded multivariate data analysis and PCA score plots for days 1 and 7, including control groups.
Fig. S4. Metabolites identified through serum metabolomics. Metabolites that showed statistically significant changes between the equidose exposures at day 1 included L-tyrosine, uric acid, taurine, pyroglutamic acid, L-carnitine, sphinganine-1-phosphate and succinic acid. Only sphinganine-1-phosphate remained perturbed between the two equidose exposures at day 7. Controls vs. 1 Gy neutron irradiated groups also showed statistical significance for L-tyrosine, L-glutamic acid, uric acid, taurine, citric acid, pyroglutamic acid and L-carnitine at day 1. Overall, a generalized downregulation of the metabolites is seen at day 1 after 1 Gy neutron irradiation, except for L-carnitine levels, which are higher.
Fig. S5. Metabolites from urine and serum metabolomics together with lipidomics, which were utilized for the construction of the ROC curves; and heatmap depiction of patterns of change between 1 Gy exposures at days 1 and 7 postirradiation.
Table S1. Chromatographic and mass spectrometry details on the experiments.
Table S2. Mean values of normalized relative abundance urine metabolomics data.
Table S3. Mean values of normalized relative abundance of serum metabolomics data.
Table S4. Mean values of normalized relative abundance of serum lipidomics data.
Table S5. AUC values of each metabolite from individual ROC curves.
Acknowledgments
This work was funded by the National Institutes of Health (National Institute of Allergy and Infectious Diseases), grant no. U19 AI067773 (PI David J. Brenner, performed as part of Columbia University Center for Medical Countermeasures against Radiation) and grant no. P30 CA051008 (PI Louis Weiner). We also thank the Lombardi Comprehensive Cancer Proteomics and Metabolomics Shared Resource (PMSR) for data acquisition, particularly Dr. Amrita Cheema and Ms. Kirandeep Gill. The content of this work is solely the responsibility of the authors and does not necessarily represent the official views of the agencies listed above.
References
- 1.Dicarlo AL, Maher C, Hick JL, Hanfling D, Dainiak N, Chao N, et al. Radiation injury after a nuclear detonation: medical consequences and the need for scarce resources allocation. Disaster Med Public Health Prep. 2011;5:S32–44. doi: 10.1001/dmp.2011.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flood AB, Ali AN, Boyle HK, Du G, Satinsky VA, Swarts SG, et al. Evaluating the special needs of the military for radiation biodosimetry for tactical warfare against deployed troops: comparing military to civilian needs for biodosimetry methods. Health Phys. 2016;111:169–82. doi: 10.1097/HP.0000000000000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.NCRP Report No. 165. Bethesda: National Council on Radiation Protection and Measurements; 2011. Responding to a radiological or nuclear terrorism incident: a guide for decision makers. [DOI] [PubMed] [Google Scholar]
- 4.Coleman CN, Koerner JF. Biodosimetry: medicine, science, and systems to support the medical decision-maker following a large scale nuclear or radiation incident. Radiat Prot Dosimetry. 2016;172:38–46. doi: 10.1093/rpd/ncw155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sullivan JM, Prasanna PG, Grace MB, Wathen LK, Wallace RL, Koerner JF, et al. Assessment of biodosimetry methods for a mass-casualty radiological incident: medical response and management considerations. Health Phys. 2013;105:540–54. doi: 10.1097/HP.0b013e31829cf221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coy SL, Cheema AK, Tyburski JB, Laiakis EC, Collins SP, Fornace A. Radiation metabolomics and its potential in biodosimetry. Int J Radiat Biol. 2011;87:802–23. doi: 10.3109/09553002.2011.556177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goudarzi M, Mak TD, Chen C, Smilenov LB, Brenner DJ, Fornace AJ. The effect of low dose rate on metabolomic response to radiation in mice. Radiat Environ Biophys. 2014;53:645–57. doi: 10.1007/s00411-014-0558-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goudarzi M, Weber WM, Mak TD, Chung J, Doyle-Eisele M, Melo DR, et al. A comprehensive metabolomic investigation in urine of mice exposed to strontium-90. Radiat Res. 2015;183:665–74. doi: 10.1667/RR14011.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goudarzi M, Weber WM, Mak TD, Chung J, Doyle-Eisele M, Melo DR, et al. Metabolomic and lipidomic analysis of serum from mice exposed to an internal emitter, cesium-137, using a shotgun LC-MS(E) approach. J Proteome Res. 2015;14:374–84. doi: 10.1021/pr500913n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson CH, Patterson AD, Krausz KW, Lanz C, Kang DW, Luecke H, et al. Radiation metabolomics. 4. UPLC-ESI-QTOFMS-Based metabolomics for urinary biomarker discovery in gamma-irradiated rats. Radiat Res. 2011;175:473–84. doi: 10.1667/RR2437.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson CH, Patterson AD, Krausz KW, Kalinich JF, Tyburski JB, Kang DW, et al. Radiation metabolomics. 5. Identification of urinary biomarkers of ionizing radiation exposure in nonhuman primates by mass spectrometry-based metabolomics. Radiat Res. 2012;178:328–40. doi: 10.1667/rr2950.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laiakis EC, Mak TD, Anizan S, Amundson SA, Barker CA, Wolden SL, et al. Development of a metabolomic radiation signature in urine from patients undergoing total body irradiation. Radiat Res. 2014;181:350–61. doi: 10.1667/RR13567.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laiakis EC, Strassburg K, Bogumil R, Lai S, Vreeken RJ, Hankemeier T, et al. Metabolic phenotyping reveals a lipid mediator response to ionizing radiation. J Proteome Res. 2014;13:4143–54. doi: 10.1021/pr5005295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laiakis EC, Strawn SJ, Brenner DJ, Fornace AJ. Assessment of saliva as a potential biofluid for biodosimetry: a pilot metabolomics study in mice. Radiat Res. 2016;186:92–7. doi: 10.1667/RR14433.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laiakis EC, Pannkuk EL, Diaz-Rubio ME, Wang YW, Mak TD, Simbulan-Rosenthal CM, et al. Implications of genotypic differences in the generation of a urinary metabolomics radiation signature. Mutat Res. 2016;788:41–9. doi: 10.1016/j.mrfmmm.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mak TD, Tyburski JB, Krausz KW, Kalinich JF, Gonzalez FJ, Fornace AJ. Exposure to ionizing radiation reveals global dose-and time-dependent changes in the urinary metabolome of rat. Metabolomics. 2015;11:1082–94. doi: 10.1007/s11306-014-0765-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Menon SS, Uppal M, Randhawa S, Cheema MS, Aghdam N, Usala RL, et al. Radiation metabolomics: current status and future directions. Front Oncol. 2016;6:20. doi: 10.3389/fonc.2016.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tyburski JB, Patterson AD, Krausz KW, Slavík J, Fornace AJ, Gonzalez FJ, et al. Radiation metabolomics. 1. Identification of minimally invasive urine biomarkers for gamma-radiation exposure in mice. Radiat Res. 2008;170:1–14. doi: 10.1667/RR1265.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tyburski JB, Patterson AD, Krausz KW, Slavík J, Fornace AJ, Gonzalez FJ, et al. Radiation metabolomics. 2. Dose- and time-dependent urinary excretion of deaminated purines and pyrimidines after sublethal gamma-radiation exposure in mice. Radiat Res. 2009;172:42–57. doi: 10.1667/RR1703.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cullings HM, Pierce DA, Kellerer AM. Accounting for neutron exposure in the Japanese atomic bomb survivors. Radiat Res. 2014;182:587–98. doi: 10.1667/RR13663.1. [DOI] [PubMed] [Google Scholar]
- 21.Sasaki MS, Nomura T, Ejima Y, Utsumi H, Endo S, Saito I, et al. Experimental derivation of relative biological effectiveness of A-bomb neutrons in Hiroshima and Nagasaki and implications for risk assessment. Radiat Res. 2008;170:101–17. doi: 10.1667/RR1249.1. [DOI] [PubMed] [Google Scholar]
- 22.Xu Y, Randers-Pehrson G, Turner HC, Marino SA, Geard CR, Brenner DJ, et al. Accelerator-based biological irradiation facility simulating neutron exposure from an improvised nuclear device. Radiat Res. 2015;184:404–10. doi: 10.1667/RR14036.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu Y, Randers-Pehrson G, Marino SA, Garty G, Harken A, Brenner DJ. Broad energy range neutron spectroscopy using a liquid scintillator and a proportional counter: application to a neutron spectrum similar to that from an improvised nuclear device. Nucl Instrum Methods Phys Res A. 2015;794:234–9. doi: 10.1016/j.nima.2015.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Egbert SD, Kerr GD, Cullings HM. DS02 fluence spectra for neutrons and gamma rays at Hiroshima and Nagasaki with fluence-to-kerma coefficients and transmission factors for sample measurements. Radiat Environ Biophys. 2007;46:311–25. doi: 10.1007/s00411-007-0120-5. [DOI] [PubMed] [Google Scholar]
- 25.Broustas CG, Xu Y, Harken AD, Garty G, Amundson SA. Comparison of gene expression response to neutron and x-ray irradiation using mouse blood. BMC Genomics. 2017;18:2. doi: 10.1186/s12864-016-3436-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Broustas CG, Xu Y, Harken AD, Chowdhury M, Garty G, Amundson SA. Impact of neutron exposure on global gene expression in a human peripheral blood model. Radiat Res. 2017 doi: 10.1667/RR0005.1. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kramer K, Li A, Madrigal J, Sanchez B, Millage K. Monte Carlo modeling of the initial radiation emitted by a nuclear device in the national capital region. Fort Belvoir, VA: Defense Threat Reduction Agency; 2013. (Report No. DTRA-TR-13-045). [Google Scholar]
- 28.Pannkuk EL, Laiakis EC, Mak TD, Astarita G, Authier S, Wong K, et al. A lipidomic and metabolomic serum signature from nonhuman primates exposed to ionizing radiation. Metabolomics. 2016;12:1–11. doi: 10.1007/s11306-016-1010-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laiakis EC, Trani D, Moon BH, Strawn SJ, Fornace AJ. Metabolomic profiling of urine samples from mice exposed to protons reveals radiation quality and dose specific differences. Radiat Res. 2015;183:382–90. doi: 10.1667/RR3967.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mak TD, Laiakis EC, Goudarzi M, Fornace AJ. MetaboLyzer: a novel statistical workflow for analyzing Postprocessed LC-MS metabolomics data. Anal Chem. 2014;86:506–13. doi: 10.1021/ac402477z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, et al. HMDB 3.0–The Human Metabolome Database in 2013. Nucleic Acids Res. 2013;41:D801–7. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42:D199–205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fahy E, Sud M, Cotter D, Subramaniam S. LIPID MAPS online tools for lipid research. Nucleic Acids Res. 2007;35:W606–12. doi: 10.1093/nar/gkm324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sud M, Fahy E, Cotter D, Brown A, Dennis EA, Glass CK, et al. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 2007;35:D527–32. doi: 10.1093/nar/gkl838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kind T, Liu KH, Lee DY, DeFelice B, Meissen JK, Fiehn O. LipidBlast in silico tandem mass spectrometry database for lipid identification. Nat Methods. 2013;10:755–8. doi: 10.1038/nmeth.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith CA, O’Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, et al. METLIN: a metabolite mass spectral database. Ther Drug Monit. 2005;27:747–51. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 38.Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0– making metabolomics more meaningful. Nucleic Acids Res. 2015;43:W251–7. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saeed A, Raouf GA, Nafee SS, Shaheen SA, Al-Hadeethi Y. Effects of very low dose fast neutrons on cell membrane and secondary protein structure in rat erythrocytes. PLoS One. 2015;10:e0139854. doi: 10.1371/journal.pone.0139854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brenner DJ, Sachs RK. Chromosomal “fingerprints” of prior exposure to densely ionizing radiation. Radiat Res. 1994;140:134–42. [PubMed] [Google Scholar]
- 41.Hande MP, Azizova TV, Geard CR, Burak LE, Mitchell CR, Khokhryakov VF, et al. Past exposure to densely ionizing radiation leaves a unique permanent signature in the genome. Am J Hum Genet. 2003;72:1162–70. doi: 10.1086/375041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laiakis EC, Hyduke DR, Fornace AJ. Comparison of mouse urinary metabolic profiles after exposure to the inflammatory stressors gamma radiation and lipopolysaccharide. Radiat Res. 2012;177:187–99. doi: 10.1667/rr2771.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ayala A, Muñoz MF, Argüelles S. Lipid peroxidation: production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxid Med Cell Longev. 2014;2014:360438. doi: 10.1155/2014/360438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Balboa MA, Balsinde J. Oxidative stress and arachidonic acid mobilization. Biochim Biophys Acta. 2006;1761:385–91. doi: 10.1016/j.bbalip.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 45.Soltan Monem A, Ali FM, Al-Thani NJ, Ali SA. Membrane solubilization in erythrocytes as a measure of radiation exposure to fast neutrons. Phys Med Biol. 1999;44:347–55. doi: 10.1088/0031-9155/44/2/004. [DOI] [PubMed] [Google Scholar]
- 46.Benderitter M, Vincent-Genod L, Pouget JP, Voisin P. The cell membrane as a biosensor of oxidative stress induced by radiation exposure: a multiparameter investigation. Radiat Res. 2003;159:471–83. doi: 10.1667/0033-7587(2003)159[0471:tcmaab]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 47.Li L, Steinauer KK, Dirks AJ, Husbeck B, Gibbs I, Knox SJ. Radiation-induced cyclooxygenase 2 up-regulation is dependent on redox status in prostate cancer cells. Radiat Res. 2003;160:617–21. doi: 10.1667/rr3076. [DOI] [PubMed] [Google Scholar]
- 48.Schaue D, Micewicz ED, Ratikan JA, Xie MW, Cheng G, Mcbride WH. Radiation and inflammation. Semin Radiat Oncol. 2015;25:4–10. doi: 10.1016/j.semradonc.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang C, Yang J, Nie J. Plasma phospholipid metabolic profiling and biomarkers of rats following radiation exposure based on liquid chromatography-mass spectrometry technique. Biomed Chromatogr. 2009;23:1079–85. doi: 10.1002/bmc.1226. [DOI] [PubMed] [Google Scholar]
- 50.Pannkuk EL, Laiakis EC, Authier S, Wong K, Fornace AJ. Targeted metabolomics of nonhuman primate serum after exposure to ionizing radiation: potential tools for high-throughput bio-dosimetry. RSC Advances. 2016;6:51192–202. doi: 10.1039/C6RA07757A. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Examples of raw chromatograms for lipidomics, serum metabolomics and urine metabolomics.
Fig. S2. Weight and creatinine levels of control and irradiated mice from each group.
Fig. S3. Expanded multivariate data analysis and PCA score plots for days 1 and 7, including control groups.
Fig. S4. Metabolites identified through serum metabolomics. Metabolites that showed statistically significant changes between the equidose exposures at day 1 included L-tyrosine, uric acid, taurine, pyroglutamic acid, L-carnitine, sphinganine-1-phosphate and succinic acid. Only sphinganine-1-phosphate remained perturbed between the two equidose exposures at day 7. Controls vs. 1 Gy neutron irradiated groups also showed statistical significance for L-tyrosine, L-glutamic acid, uric acid, taurine, citric acid, pyroglutamic acid and L-carnitine at day 1. Overall, a generalized downregulation of the metabolites is seen at day 1 after 1 Gy neutron irradiation, except for L-carnitine levels, which are higher.
Fig. S5. Metabolites from urine and serum metabolomics together with lipidomics, which were utilized for the construction of the ROC curves; and heatmap depiction of patterns of change between 1 Gy exposures at days 1 and 7 postirradiation.
Table S1. Chromatographic and mass spectrometry details on the experiments.
Table S2. Mean values of normalized relative abundance urine metabolomics data.
Table S3. Mean values of normalized relative abundance of serum metabolomics data.
Table S4. Mean values of normalized relative abundance of serum lipidomics data.
Table S5. AUC values of each metabolite from individual ROC curves.


