Fig. 6.
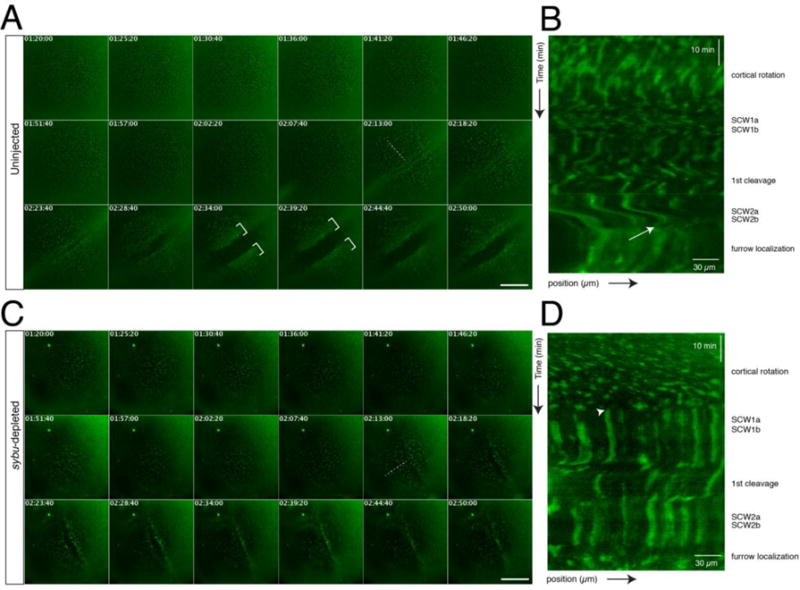
Time lapse imaging of germ plasm aggregation during early development in host-transfer fertilized eggs. (A) Representative time-lapse imaging of a zygote obtained by the host-transfer procedure, stained with DiOC6(3). Panels in (A) show selected frames at ~5-minute intervals from Supplementary Video 1 (170 minute time-lapse, frame rate = 20 seconds). In (A, C), elapsed time from in vitro fertilization is shown at the top of each frame (in hr:min:sec); dotted lines indicate the lines used for kymograph profiles; brackets indicate areas of clustered germ plasm; scale bars = 85 μm. (B) Representative kymograph analysis of an uninjected control embryo. The approximate timing of post-fertilization events is shown on the right. The white arrow indicates the beginning of aggregation into larger areas at the tail end of SCW2a/b. The position arrow indicates distance from the vegetal pole (bottom left corner). (C) Representative time-lapse imaging of a sybu-depleted zygote obtained by the host-transfer procedure, stained with DiOC6(3) (labels as in A). Panels show selected frames at ~5-minute intervals from Supplementary Video 2. (D) Representative kymograph analysis of a sybu-depleted embryo. Germ plasm is not disturbed by SCWs (arrowhead) and fails to form large continuous aggregates at the cleavage furrows.
