Abstract
Hepatic fibrosis represents a pathological wound healing and tissue repair process triggered in response to chronic liver injury. A heterogeneous population of activated non-parenchymal liver cells, known as liver myofibroblasts, functions as the effector cells in hepatic fibrosis. Upon activation, liver myofibroblasts become fibrogenic, acquiring contractile properties and increasing collagen production capacity, while developing enhanced sensitivity to endogenous molecules and factors released in the local microenvironment. Hepatic extracellular adenosine is a bioactive small molecule, increasingly recognized as an important regulator of liver myofibroblast functions, and an important mediator in the pathogenesis of liver fibrosis overall. Remarkably, ecto-5′-nucleotidase/Nt5e/Cd73 enzyme, which accounts for the dominant adenosine-generating activity in the extracellular medium, is expressed by activated liver myofibroblasts. However, the molecular signals regulating Nt5e gene expression in liver myofibroblasts remain poorly understood. Here, we show that activated mouse liver myofibroblasts express Nt5e gene products and characterize the putative Nt5e minimal promoter in the mouse species. We describe the existence of an enhancer sequence upstream of the mouse Nt5e minimal promoter and establish that the mouse Nt5e minimal promoter transcriptional activity is negatively regulated by an Elf2-like Ets-related transcription factor in activated mouse liver myofibroblasts.
Keywords: Ecto-5′-nucleotidase (Nt5e), Cluster of differentiation 73 (Cd73), Surface enzyme, Adenosine, Transcriptional regulation, Promoter, ETS transcription factor family, E74-like factor 2/new ETS-related factor (Elf2/Nerf), Cloning, Electric mobility shift assay
Introduction
Hepatic fibrosis, evolving to cirrhosis, is a hallmark feature of almost all chronic liver diseases. This tissue repair and wound healing process is triggered in response to tissue/cell injury or exposure to various insults [1]. It is well known that liver myofibroblasts (MF) are the primary effector cells responsible for extracellular matrix production and scar formation observed during hepatic fibrosis, in both clinical and experimental settings [2]. Essentially absent from the healthy tissue, MF rapidly accumulate within the injured liver [3]. Liver MF may derive, upon cell activation, from a variety of sources, including intrahepatic non-parenchymal cells, such as hepatic stellate cells, periportal and perivascular fibroblasts, mesothelial cells, and extrahepatic cells, such as bone marrow-derived fibrocytes [4]. However, hepatic stellate cells (HSC) and portal fibroblasts (PF) are by far the best-characterized sources of liver MF [5]. As the major liver fibrogenic cells implicated in tissue repair and wound healing response, liver MF represent excellent targets for anti-fibrotic therapies and treatment of chronic liver diseases.
During liver fibrosis, MF functionally contribute to critical pathophysiological processes such as local inflammation, vascular remodeling, and tissue regeneration, which are regulated by a number of small signaling molecules including extracellular ATP nucleotide and its degradation end product adenosine nucleoside [6]. Particularly, extracellular adenosine nucleoside can induce signal transduction through four transmembrane receptors in liver MF and modulate various cell functions/properties that impact the progression of hepatic fibrosis, such as matrix protein production, cytokine release, cell proliferation, and cell motility [7, 8]. Fittingly, experimental liver fibrosis models in rodents using gene silencing or pharmacological inhibition approaches targeting adenosine receptors have demonstrated the biological relevance of extracellular adenosine as a critical regulator of the hepatic fibrogenesis process [9–11]. Although most evidence points towards pro-fibrotic actions for adenosine, it can also exert anti-fibrotic effects in the liver [9] and other organs [12].
Within the liver, the primary determinant of tissue adenosine levels is the cell surface Cd73/Nt5e/ecto-5′-nucleotidase enzyme [13]. In the healthy liver, Cd73 expression is mostly found in parenchyma, endothelium, and both quiescent hepatic stellate cells, and portal cells [14]. In the fibrosing liver, it is redistributed to, and transcriptionally regulated, the liver MF deriving from activated HSC and PF of murine and human origins [15–18]. For instance, Sp1/Smad transcription factors regulate Cd73 gene expression in rat HSC-derived liver MF [15]. Moreover, other transcription factors that have functional relevance in the setting of chronic liver injury, such as Hif1α [19], Stat3 [20], and Tgf-β [21] have been shown to regulate Cd73 gene expression in other cell types. In addition, Cd73 has been shown to participate in and influence fibrosis progression experimentally [14, 22]. Taken together, these data suggest that regulation of liver Cd73 expression is critical in the pathogenesis of hepatic fibrosis. Therefore, to further elucidate the genetic mechanisms regulating Cd73 expression in liver MF, we sought to clone and characterize a mouse genomic DNA corresponding to the putative Nt5e gene promoter.
Experimental procedures
Materials and reagents
Cell culture reagents and media were obtained from Life Technologies (Life Technologies, Carlsbad, CA), Fisher Scientific (Fisher Scientific, Pittsburgh, PA), and Thermo Scientific (Thermo Scientific, Rockland, MA). Molecular biology and SDS-PAGE reagents and kits were obtained from Qiagen (Qiagen, Valencia, CA), Bio-Rad Laboratories (Bio-Rad Laboratories, Hercules, CA), and Life Technologies. Restriction enzymes and T4 DNA ligase were obtained from New England Biolabs (Ipswich, MA).
Animal care
All mouse experiments were performed in accordance with regulations approved by the University of Arkansas for Medical Sciences Institutional Animal Care and Use Committee. Male C57Bl6 mice (6–8 weeks, n = 5 per group) were purchased from Charles River Laboratories (Redfield, AR) and subjected to experimental liver fibrosis induced by carbon tetrachloride intoxication (0.5 ml/kg diluted in sterile olive oil intraperitoneally, 9 injections total) for 3 weeks, as previously described [22].
Cell culture
Immortalized NIH3T3 mouse embryonic fibroblasts [24], JS1 mouse myofibroblastic hepatic stellate cells [25], and Col-GFP mouse myofibroblastic hepatic stellate cells [26] were grown in DMEM supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin antibiotics as described previously. All cells were maintained at 37 °C, under 95% air–5% CO2.
Primary cell isolation and culture
Primary HSC were isolated from mouse livers as previously described [27]. Briefly, hepatocyte and non-parenchymal cell fractions were obtained by in situ pronase/collagenase perfusion of livers. Mouse non-parenchymal cells were subsequently used for isolation of HSC by density gradient centrifugation [28]. The resulting cell suspension were plated onto tissue culture plastic dishes in DMEM/F-12 containing 10% FBS and antibiotics. Primary mouse HSC were used before day 3 (quiescent) and past day 4 (myofibroblastic/activated) after plating, as previously described [28]. All cells were maintained at 37 °C, under 95% air–5% CO2.
RT-PCR
Total RNA was isolated from cultured mouse JS1, Col-GFP, and primary mouse HSC cells using the RNeasy Plus Kit (Qiagen). Each RNA sample (1 μg) was digested with DNase 1 enzyme (Ambion, Life Technologies) to remove any genomic DNA contamination and reverse-transcribed using the iScript RT Supermix (Bio-Rad). Each RT reaction product (1 μl of 10 fold-dilution) was further used for PCR amplifications. Semi-quantitative PCR amplification was performed using the TopTaq Master Mix Kit (Qiagen) with the following protocol for the PCR reactions: Initialization at 94 °C for 2 min followed by 35 cycles of 30 s denaturation at 94 °C, 30 s annealing at 60 °C, and 30 s elongation at 72 °C; the amplification was then completed with a 10-min final elongation at 72 °C using an S1000 Thermo Cycler (Bio-Rad). The primer sequences used for the semi-quantitative RT-PCR reactions are listed in Table 1. Amplification products were visualized on 3% agarose gels via ethidium bromide staining. Real-time PCR amplification was performed using the SsoAdvanced Universal Real-Time PCR Supermix (Bio-Rad) and probes for Nt5e (ecto-5′-nucleotidase, IDT#Mm.PT.56a.12096204), Elf2 (E74-like factor 2, IDT#Mm.PT.58.9437524), and housekeeping Gapdh (glyceraldehyde 3-phosphate dehydrogenase, IDT#Mm.PT.39a.1) genes in mouse species. The following PCR protocol was used: initial denaturation step at 95 °C for 3 min, followed 40 repetitions of a 2-step cycling program consisting of 10 s at 95 °C, 30 s at 57 °C. Relative fold change in mRNA expression was estimated using the 2(−Delta Delta C(T)) method [29].
Table 1.
The primer sequences used for the PCR amplification reactions
| Name | Sequence | Location (relative to ATG) | Experiment |
|---|---|---|---|
| pmCd73-R | AGATCTCTGGCTAGAGCGCGTTGAG | −20 to −3 | Promoter cloning |
| pmCd73-F1 | GGTACCAGAGATTTGCTTGCCTTTGC | −2069 to −2050 | Promoter cloning |
| pmCd73-F2 | GGTACCATGCAGACCTGGTGCATTCT | −1010 to −991 | Promoter cloning |
| pmCd73-F3 | GGTACCGGTTGCGTCCTCAAACCTAA | −567 to −548 | Promoter cloning |
| pmCd73-F4 | GGTACCCAGCCTCTCGGACCTGCT | −275 to −258 | Promoter cloning |
| pmCd73-F5 | GGTACCGTGACTCGGGGAGTGTGTCT | −162 to −143 | Promoter cloning |
| pmCd73-F6 | GGTACCTCTACTCCGCAGTTTAGTAGAGG | −87 to −65 | Promoter cloning |
| pmCd73-R-ENH | AGATCTGCACCAGGTCTGCATTGTT | −1014 to −996 | Enhancer cloning |
| pmCd73-L-ENH1074 | GGTACCAGAGATTTGCTTGCCTTTGC | −2069 to −2050 | Enhancer cloning |
| pmCd73-L-ENH511 | GGTACCATGCAATGTTGACTGCCAGA | −1506 to −1487 | Enhancer cloning |
| pmCd73-R’ | AAGCTTCTGGCTAGAGCGCGTTGAG | −20 to −3 | Enhancer cloning |
| pmCd73-F5’ | AGATCTGTGACTCGGGGAGTGTGTCT | −162 to −143 | Enhancer cloning |
| mutElf2 sense | GGGCCTGGGGC A GGAAG T G A CGGGTCTGTCC | −132 to −102 | Mutagenesis, EMSA probe |
| mutElf2 antisense | GGACAGACCCG T C A CTTCC T GCCCCAGGCCC | −132 to −102 | Mutagenesis, EMSA probe |
| mElf2 L | GTCCTGGCAATCGTCTGTGA | N/A | semi-quantitative PCR |
| mElf2 R | TGGGAACTGGCTCCACAATC | N/A | semi-quantitative PCR |
| mCd73 L | ACTCCACCAAGTGCCTCAAC | N/A | semi-quantitative PCR |
| mCd73 R | GGGATCAATCAGTCCTTCCA | N/A | semi-quantitative PCR |
| mGapdh L | TTGTGCAGTGCCAGCCTC | N/A | semi-quantitative PCR |
| mGapdh R | CTGGAAGATGGTGATGGGCT | N/A | semi-quantitative PCR |
| mCd73-elf2 L1 | AGGGCCTGGGGCTGGAAGAGGCGGGTCTGT | −131 to −100 | EMSA probe |
| mCd73-elf2 R1 | ACAGACCCGCCTCTTCCAGCCCCAGGCCCT | −131 to −100 | EMSA probe |
Bold emphasis indicates added restriction enzyme site(s). Underlined letters indicate base pair change(s)
Immunohistochemistry and immunofluorescence
For immunohistochemistry, liver sections from paraffin-embedded tissues were prepared at 5-μm thickness using a routine procedure at the University of Arkansas for Medical Sciences Experimental Pathology Core Laboratory. Serial sections were used for Masson’s Trichrome staining method to identify interstitial collagen fibrils with blue coloring. Another set of sections were used for immunostaining method using the standard Vectastain Elite ABC HRP kit according to the manufacturer’s instructions (#PK-6100, Vector Laboratories, Burlingame, CA) to monitor the liver distribution of mouse alpha smooth muscle actin (αSMA), Cd73, and Elf2 proteins with the following primary antibodies: rabbit polyclonal anti-αSMA (ab#5694, Abcam, Cambridge, MA), rabbit monoclonal anti-mouse/rat/human Cd73 (clone D7F9A, #13160S, Cell Signaling Technology, Danvers, MA), and rabbit polyclonal anti-mouse/rat/human Elf2 (#NBP1–84770, Novus Biologicals, Littleton, CO) antibodies, and a secondary donkey biotinylated anti-rabbit IgG antibody (Affinipure #711-065-152, Jackson ImmunoResearch Laboratories, West Grove, PA). Mowiol-based mountant was then added to stained tissue sections. Bright-field microscopy images were acquired using an Olympus BX-51 microscope (Olympus America, Melville, NY). For immunofluorescence, primary culture-activated HSC were plated and grown overnight on coverslips. The following day, cells were fixed with neutral (pH = 7.2) 4% paraformaldehyde solution (diluted in 1× Phosphate-Buffered Saline, PBS) for 20 min, washed in 1× PBS, and further permeabilized with Triton X-100 0.1% solution (diluted in 1× PBS) for 10 min, all steps at room temperature. After several washes in 1× PBS, coverslips were incubated with a 7% goat serum (Life Technologies), 0.5% bovine serum albumin (Fisher Scientific) blocking solution (diluted in 1× PBS) at room temperature for 1 h, and then with rabbit polyclonal anti-αSMA (ab#5694, Abcam), rabbit polyclonal anti-mouse/rat/human collagen type 1 (#234167, EMD Millipore, Billerica, MA), rabbit polyclonal anti-Pdgfrβ (958) (#sc-432, Santa Cruz BioTechnology, Dallas TX), and rabbit monoclonal anti-mouse/rat/human Cd73 (clone H300, #sc-25603, Santa Cruz BioTechnology) at 4 °C overnight. After several washes in 1× PBS, coverslips were further incubated with a goat Alexa488-conjugated anti-rabbit IgG antibody (diluted in blocking solution). After several washes in 1× PBS, DAPI-supplemented Prolong Diamond anti-fade mountant (Life Technologies) was added to coverslips. As negative controls, primary antibody or labeling probe was omitted, with no staining occurring (not shown). Confocal fluorescence microscopy images were acquired using Zeiss LSM 510 Meta imaging system (Zeiss Laboratories, White Plains, NY).
Plasmids
The genomic DNA sequence corresponding to the 5′ untranslated region of mouse Nt5e gene (NCBI Gene ID 23959) was extracted from the NCBI/GenBank database to determine the location of ATG start codon (defined as +1 position). To analyze the promoter activity of the 5′ untranslated region of mouse Nt5e gene, six separate fragments of the promoter region with length varying between 2067 (−2069 to −3 position relative to start codon) and 85 (−87 to −3) base pairs (bp) were initially generated using a PCR-based cloning approach. An additional restriction enzyme site was added to each forward (KpnI) and reverse (BglII) primer (primer sequences and positions are listed in Table 1). For PCR amplification of the longest fragment (2067 bp) that was hypothesized to contain the complete promoter, 100 ng of commercially available mouse genomic DNA (Clonetch, Mountain View, CA) was used as template with the primers pmCd73-F1 and pmCd73-R. Approximately 10 ng of the first plasmid DNA was then used as template for PCR amplification of all other promoter fragments, with five different forward primers (pmCd73-F2 to pmCd73-F6) and the same reverse primer (pmCd73-R). All PCR amplifications were done with Expand High-Fidelity PCR System (Roche Biosciences) for maximum of fidelity of the elongation, as follows: 95 °C for 2 min, 30 cycles of 95 °C for 30 s (denaturation), 62 °C for 30 s (annealing), and 72 °C for 2 min 30 s (elongation), and completed by a 7-min incubation at 72 °C. Amplification products were ligated into the KpnI/BglII cloning site in the Luciferase reporter gene pGL3-Basic vector (Promega, Madison, WI) to generate pmCd73 F1- to F6-Luc constructs.
To characterize a putative 1074-bp-long regulatory sequence (−2069 to −996) contained in the longest fragment (−2069 to −3) of the promoter region (F1-Luc construct), one fragment of the promoter region with length of 160 bp (−162 to −3) was generated by PCR. An additional restriction enzyme site was added to each forward (BglII) and reverse (HindIII) primer (primer sequences and positions are listed in Table 1). Approximately 10 ng of the F5-Luc plasmid DNA (described above) was used as template for PCR amplification with forward (pmCd73-F5′) and reverse (pmCd73-R′) primers and Expand High-Fidelity PCR System (Roche Biosciences) for maximum of fidelity of the elongation as described above, except for elongation time that was 30 s. Amplification products were ligated into the BglII/HindIII cloning site in the Luciferase reporter gene pGL3-Basic vector to generate the ENH-F5-Luc construct. Next, two separate fragments of the promoter region 1074 bp (−2069 to −996) and 511 bp (−1506 to −996) were generated by PCR. An additional restriction enzyme site was added to each forward (KpnI) and reverse (BglII) primer (primer sequences and positions are listed in Table 1). Approximately 10 ng of the F1-Luc plasmid DNA (described above) was used as template for PCR amplification of all promoter fragments with two different forward primers (pmCd73-L-ENH1074 and pmCd73-L-ENH511) and the same reverse primer (pmCd73-R-ENH) and primer sets corresponding to four contiguous portions of the analyzed 564-bp-long regulatory sequence. All PCR amplifications were done with Expand High-Fidelity PCR System (Roche Biosciences) for maximum of fidelity of the elongation, as described above, except for elongation time that was 1 min and 10 s. Amplification products were ligated into the KpnI/BglII cloning site in the Luciferase reporter gene pGL3-Basic vector to generate ENH1074- and ENH511-F5-Luc constructs. Plasmid nucleotide sequences were confirmed by automated sequencing at the UAMS Sequencing Core Facility using a 3130XL Genetic Analyzer (Applied Biosystems, Foster City, CA).
Site-directed mutagenesis
A putative promoter responsive element for mouse Elf2 transcription factor was identified using the bioinformatics software tool MatInspector (Genomatix GmbH, Munich, Germany), and a fragment lacking such element was generated by site-directed mutagenesis. Site-directed mutagenesis was performed using binding site-specific primer sets and the QuikChange II XL kit (Stratagene, La Jolla, CA). PCR amplification of mutants was performed using PfuUltra High Fidelity DNA polymerase (Stratagene) under the following parameters: 95 °C for 1 min, followed by 18 cycles of 95 °C for 50 s, 60 °C for 50 s, 68 °C for 6 min, and then 68 °C for 7 min. Sequence mutation was confirmed by automated sequencing before use.
DNA transfection
NIH3T3 cells were split into 24-well plates (0.5 × 105 /well) on the day before transfection. Plasmid DNAs (300 ng of each Firefly Luciferase reporter vector and 10 ng of Renilla luciferase vector) were mixed with Fugene6 transfection reagent (1 μl; Roche) diluted in Opti-MEMI (total volume 20 μl/well). The mixture was incubated for 20–25 min at room temperature and then added to cells in a stepwise fashion. Transfected cells were allowed to grow for 48 h before testing transgene expression. Transfections for every construct were performed independently in triplicate and repeated at least three times.
Luciferase activity assays
Changes in firefly luciferase activity of transfected NIH3T3 cells were normalized to Renilla luciferase activity using the Dual-Luciferase Reporter assay system (Promega) and detected using a Synergy HT multidetection microplate reader (BioTek, Winooski, VT).
Electromobility shift assays
Nuclear extracts from JS1 and Col-GFP cells were prepared using the Subcellular Protein Fractionation Kit (#NBP2–47659, Novus Biologicals) according to the manufacturer’s instructions. DNA-binding assays were performed with nuclear extracts using the DIG Gel shift 2nd generation kit (Roche Biosciences) according to the manufacturer’s instructions. The DIG-labeled double-stranded oligonucleotides used as probes are described in Table 1. For competition experiments, a 125-fold molar excess of unlabeled double-stranded oligonucleotides was added to the binding reaction. Assays were performed independently using at minimum two preparations of nuclear extracts for each cell line.
Statistical analysis
One-way ANOVA followed by Bonferroni’s post hoc Multiple Comparison test and two-tailed unpaired t test were performed using GraphPad Prism v5.00, GraphPad Software (San Diego, CA, USA, www.graphpad.com). P value (p) was considered significant when *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Results
We have previously cloned and characterized the rat Nt5e promoter and shown that Nt5e gene expression is transcriptionally regulated in activated liver myofibroblasts during fibrosis in the rat species [15]. Here, we use a similar approach to identify Nt5e gene promoter in the mouse species and study the regulation of its transcriptional activity in activated mouse liver myofibroblasts.
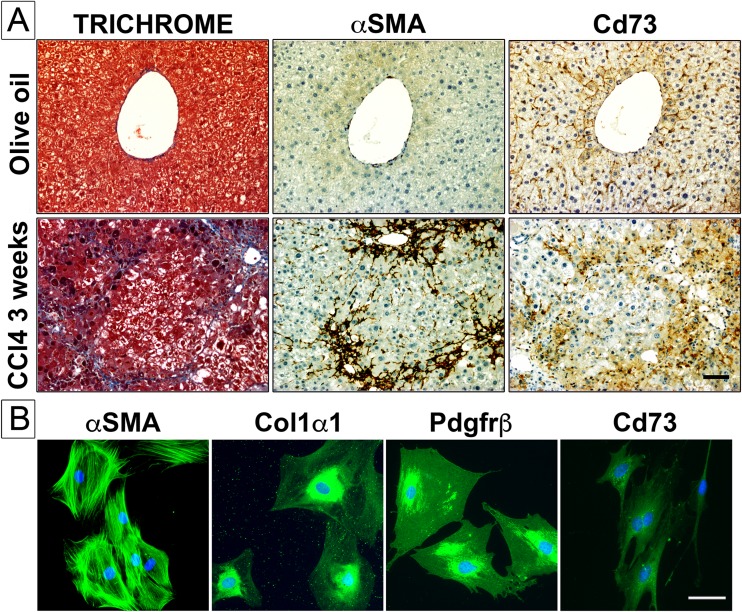
Experimental fibrosis was induced in livers of wild-type mice subjected to carbon tetrachloride (CCl4) intoxication for 3 weeks, and protein expression of classical activated liver myofibroblast marker alpha smooth muscle actin (αSMA) and purinergic cell-surface marker Cd73 was monitored and analyzed in livers of normal and fibrotic animals (Fig. 1a). In the CCl4 model, histological examination of serial liver tissue sections from olive oil vehicle-injected animals shows, as expected, the typical organization of normal liver tissue, with obvious expression of collagen in perivascular areas (Fig. 1a, TRICHROME, olive oil), modest expression of alpha smooth muscle actin that is restricted to portal areas surrounding arteries and veins (Fig. 1a, αSMA, olive oil), while expression of Cd73 is largely observed in hepatocytes and circulating mesenchymal cells (likely hepatic stellate cells) (Fig. 1a, Cd73, olive oil). In contrast, in CCl4-intoxicated animals, induction of fibrosis is clearly observed in injured liver areas, as evidenced by important structural changes in hepatic tissue architecture, prominent accumulation of extracellular matrix deposition, mainly fibrillar collagen (Fig. 1a, TRICHROME, CCl4 3 weeks), and expression of both contractile α-smooth muscle actin (Fig. 1a, αSMA, CCl4 3 weeks) and Cd73 proteins in a number of activated liver myofibroblasts (Fig. 1a, Cd73, CCl4 3 weeks). Finally, expression of Cd73 protein, along the one of classical αSMA, collagen 1 alpha 1 (Col1α1), and beta-type platelet-derived growth factor receptor (Pdgfrβ) markers, was detected in primary isolated mouse HSC upon culture activation (Fig. 1b). Taken together, these data demonstrate that Cd73 protein is expressed in activated mouse liver myofibroblasts during hepatic fibrosis, as recently reported [18].
Fig. 1.
a Experimental liver fibrosis was induced by carbon tetrachloride (CCl4) intoxication for 3 weeks. In serial sections of livers from control (olive oil-injected) animals, the typical distribution of collagen fibrils (perivascular areas), αSMA (blood vessels), and Cd73 (parenchymal, sinusoidal areas) is observed. In serial sections of livers from fibrotic (CCl4-intoxicated) animals, important accumulation of collagen fibrils is seen around nodules of parenchymal cells, along with αSMA and Cd73 proteins. Scale bar 200 μm. b αSMA, Col1α1, Pdgfrβ, and Cd73 protein expression and distribution (green color) is assessed by immunofluorescence staining, in primary mouse hepatic stellate cells upon culture activation. Culture-activated αSMA+ Col1α1+ Pdgfrβ+ are positive for Cd73 protein expression. DAPI (blue color) was used as nuclear stain. Scale bar 50 μm
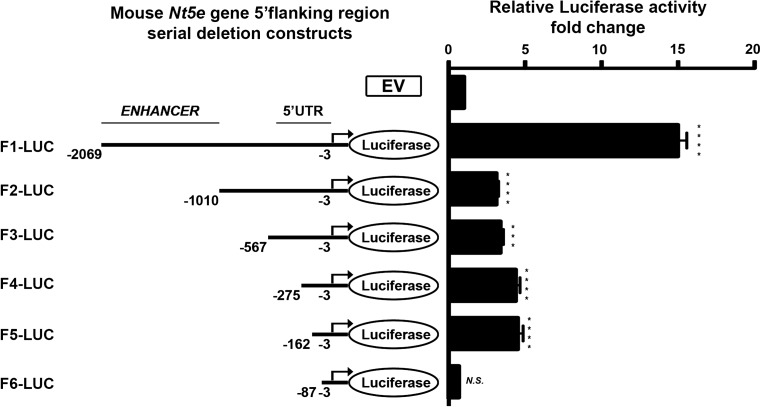
To better understand how Nt5e gene expression is regulated in mouse liver myofibroblasts, we sought to clone and characterize the mouse Nt5e promoter. The ATG start codon of the mouse Nt5e gene was extracted from the reference NCBI database (Fig. 2). A 2067-bp-long mouse genomic DNA sequence corresponding to the putative Nt5e gene promoter was cloned into a luciferase reporter plasmid (F1-Luc, −2069 to −3 bp relative to ATG start codon) and used as template for the subsequent generation of serial truncation luciferase constructs (F2- to F6-Luc). The transcriptional activity of each construct was studied by a dual-luciferase assay upon transfection in mouse NIH3T3 fibroblasts that are deficient for Nt5e gene expression and can be transfected with high efficiency (Fig. 3). Of all serial deletion constructs generated, the F1-Luc construct (−2069 to −3 bp) displayed the highest luciferase activity relative to empty vector control plasmid (F1-Luc, 14.99 ± 0.59 vs. EV, 1 ± 0.04, ****p < 0.0001), which was significantly elevated compared with shorter F2- to F6-Luc constructs (F2-Luc, 3.122 ± 0.14; F3-Luc, 3.389 ± 0.19; F4-Luc, 4.391 ± 0.28; F5-Luc, 4.532 ± 0.35; F6-Luc, 0.6554 ± 0.05, ****p < 0.0001), suggesting the existence of enhancer element(s) in the cloned putative mouse Nt5e promoter sequence. There was no significant change in luciferase activity when comparing F2- to F4-Luc constructs altogether. Furthermore, the luciferase activity of F5-Luc construct was significantly higher than the one of F2-Luc construct (F2-Luc, 3.122 ± 0.14 vs. F5-Luc, 4.532 ± 0.3, *p < 0.05). The luciferase activity of F6-Luc construct was almost completely abolished with levels similar to the ones measured for the control empty vector (F6-Luc, 0.6554 ± 0.05 vs. EV, 1 ± 0.04, NS), indicating the F5-Luc (−162 to −3 bp) construct as the minimal promoter required for the transcriptional activity of the mouse Nt5e gene, following transient transfection in NIH3T3 fibroblasts.
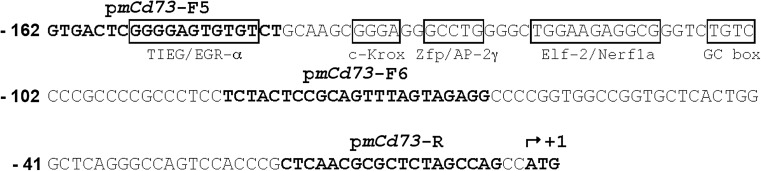
Fig. 2.
The putative mouse Nt5e minimal promoter nucleotide sequence was extracted from NCBI/GenBank database, and putative binding motifs for transcription factors were identified and scored using bioinformatics analysis. An upward arrow at the nucleotide +1 indicates the adenine of the translation initiation site (ATG). The pmCd73-F5, pmCd73-F6, and pmCd73-R primer sequences are indicated in bold type. Black empty boxes indicate predicted transcription factor binding sites
Fig. 3.
Serial truncations of mouse wild-type Nt5e promoter-luciferase constructs were transiently expressed in mouse NIH3T3 fibroblasts. NIH3T3 cells were transfected with baseline control pGL3 plasmid (empty vector, EV) or serial deletions (F2- to F6-Luc) of the putative mouse Nt5e promoter (F1 or nucleotide sequence spanning −2069 to −3 bp from ATG), and luciferase activity was determined relatively to that of EV. Maximal luciferase activity was measured for F1-Luc construct, which was significantly higher than the ones of F2- to F6-Luc constructs. The measured luciferase activity noticeably decreased between F1- and F2-Luc constructs, suggesting that F1-Luc construct contains a potential enhancer sequence, which spans approximately 1000 bp (−2069 to −996 bp) and is absent in the shorter F2-Luc construct. When compared, luciferase activities of F3- to F5-Luc constructs were not meaningfully different. Also, luciferase activity of F6-Luc construct was notably reduced relatively to that of F5-Luc construct and essentially similar to that of EV, indicating that F5-Luc construct (−162 to −3 bp) represents the minimal promoter required for mouse Nt5e gene expression (data are representative of, at least, three independent experiments)
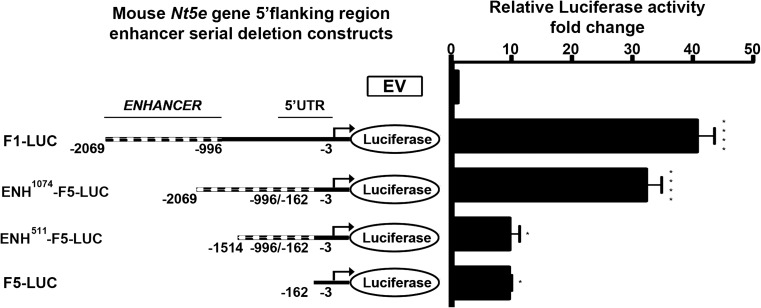
We next attempted to characterize the putative “enhancer” (−2069 to −996 bp) sequence entirely spanning the genomic DNA region present F1-Luc construct, but deleted in F2-Luc construct. The full (−2069 to −996 bp) or a short portion (−1514 to −996 bp) of the putative enhancer sequence was cloned immediately adjacent to the identified putative mouse Nt5e promoter corresponding to F5-Luc construct to generate ENH1074- and ENH511-F5-Luc constructs. Wild-type and enhancer-containing F5-Luc constructs were transfected in NIH3T3 cells, followed by luciferase activity measurement using a dual-luciferase assay (Fig. 4). Luciferase activity assays showed that the ENH1074-F5-Luc construct containing the full putative enhancer (−2069 to −996 bp) sequence has the ability to enhance the luciferase activity of the identified putative mouse Nt5e promoter, i.e., F5-Luc construct (ENH1074-F5-Luc, 32.18 ± 2.61 vs. F5-Luc, 9.389 ± 0.51, ****p < 0.0001). In contrast, the ENH511-F5-Luc construct containing a shorter portion of the putative enhancer sequence (−1514 to −996 bp) displays luciferase activity levels similar to the one measured for the F5-Luc construct (ENH511-F5-Luc, 9.467 ± 1.71 vs. F5-Luc, 9.389 ± 0.51, NS). These data suggest that key elements responsible for that enhancer activity are contained, at least in part, in the genomic DNA region corresponding to −2069 to −1514 bp relative to ATG star codon. Of note, in additional control experiments, versions of ENH1074- and ENH511-F5-Luc constructs lacking the sequence (−162 to −3 bp) corresponding to the minimal putative mouse Nt5e promoter did not display detectable luciferase activity, indicating that the putative enhancer sequence cannot act as a promoter per se (not shown).
Fig. 4.
A putative regulatory genomic DNA sequence (nucleotides −2069 to −996 bp relative to mouse Nt5e gene start codon) was cloned upstream of the mouse wild-type Nt5e promoter-luciferase (F5) construct (nucleotides −162 to −3 bp) before transient expression in mouse NIH3T3 fibroblasts. The luciferase activity of a F5-Luc construct is enhanced when 5′flanked by a genomic DNA sequence corresponding to nucleotides −2069 to −996 bp relative to mouse Nt5e gene start codon. This increase in luciferase activity is suppressed in a F5-Luc construct when 5′flanked by a genomic DNA sequence corresponding to nucleotides −1514 to −996 bp relative to mouse Nt5e gene start codon (data are representative of, at least, three independent experiments)
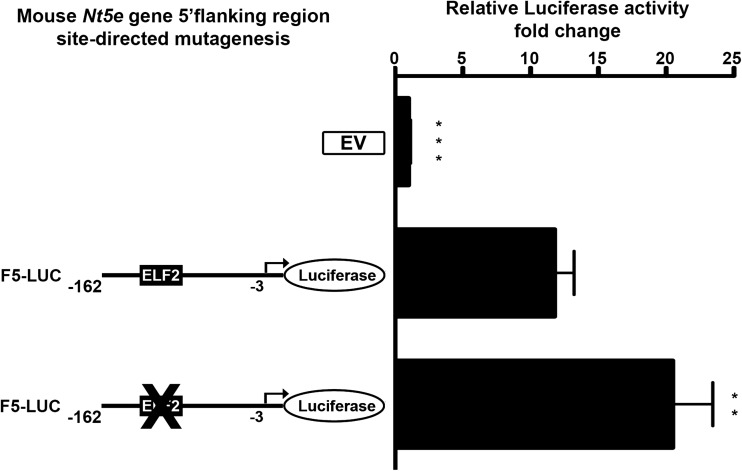
We further performed a detailed analysis of the putative mouse Nt5e promoter elements to better understand the mechanisms involved in the regulation of its transcriptional activity. In silico analysis of the putative mouse Cd73 minimal promoter F5 (−162 to −3 bp) construct revealed the presence of a response element (−121 to −113 bp) for E74-like factor 2/new E26 transformation-specific (Ets)-related factor (Elf2/Nerf) among other potential transcription factors identified (Fig. 2). Of note, Elf2 transcription factor is known to be involved the maturation and differentiation process in immune cells by gene regulation [30]. We evaluated the impact of site-directed mutagenesis targeting Elf2 transcription factor binding site on the luciferase activity of F5-Luc construct. Wild-type and mutant F5-Luc constructs were transfected in NIH3T3 cells, followed by luciferase activity measurement using a dual-luciferase assay (Fig. 5). We found that mutation of Elf2 responsive element alone was sufficient to significantly increase the F5-Luc construct luciferase activity (F5-Luc, 11.79 ± 1.41 vs. mut Elf2-F5-Luc, 20.48 ± 2.96, **p < 0.01). These data indicate that repression of mouse Nt5e promoter activity is likely mediated by transcription factor(s) acting at the identified Elf2 site in NIH3T3 fibroblasts. Similar site-directed mutagenesis experiments testing the impact of binding motifs for TGF-beta-inducible early-response gene (Tieg) [31], zinc finger, and BTB domain containing 7B (Zbtb7b/c-krox) [32] and Activator protein 2γ (Ap-2γ) [33] transcription factors identified by in silico analysis did not reveal the presence of other biologically relevant regulatory elements (not shown).
Fig. 5.
Site-directed mutagenesis of the mouse wild-type Nt5e promoter-luciferase (F5) construct (−162 to −3 bp relative to mouse Nt5e gene start codon) was performed before transient expression in mouse NIH3T3 fibroblasts. The luciferase activity of a F5-Luc construct which contains a mutated mouse Elf2 transcription factor binding site was significantly higher than the one measured for the un-mutated F5-Luc construct, indicating that Elf2-like transcription factor acts as a potential repressor of mouse Nt5e gene expression (data are representative of, at least, three independent experiments)
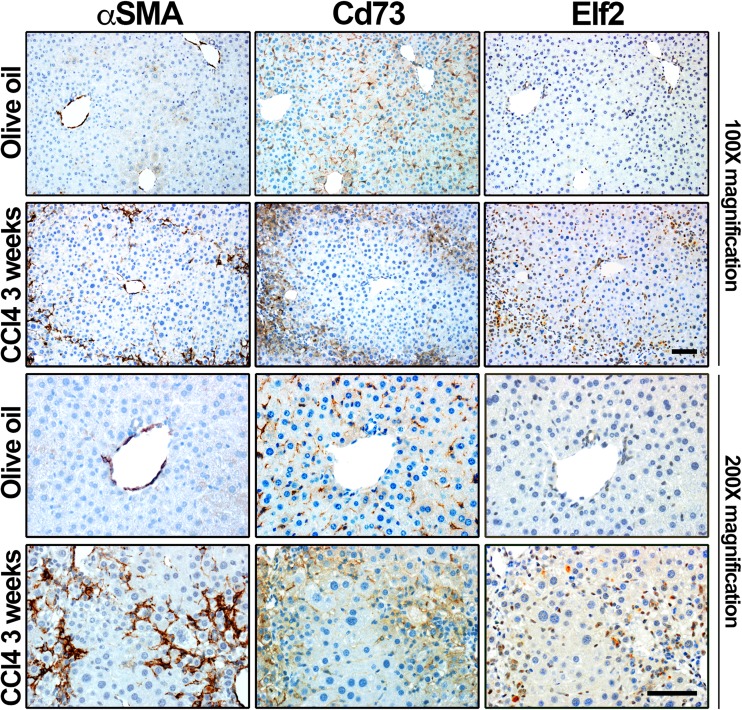
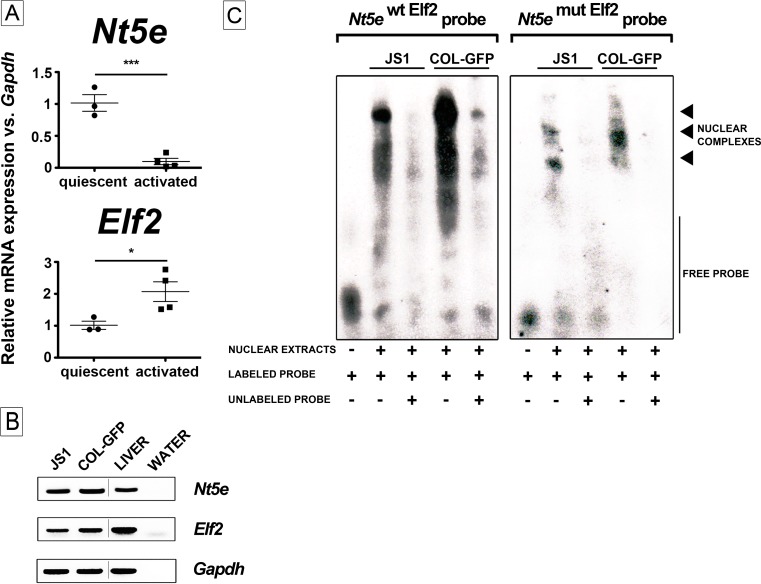
We next monitored and analyzed the expression of Elf2 protein in CCl4-induced fibrotic mouse livers (Fig. 6). Histological examination shows that a faint nuclear Elf2 staining is mostly observed in biliary epithelial cells and mesenchymal cells in vehicle-injected animals (Fig. 6a, olive oil, Elf2); it clearly redistributed to the nuclei of myofibroblasts present within fibrotic liver tissue areas in CCl4-intoxicated animals (Fig. 6, CCl4 3 weeks, Elf2). Low power field images show Elf2 staining along those of αSMA and Cd73 clearly outlines fibrotic nodules in CCl4-intoxicated animals (Fig. 6, CCl4 3 weeks, Elf2). In addition, Elf2 antibody did not appear to stain hepatocytes found in damaged areas surrounding scarring liver tissue. In summary, these data demonstrate that Elf2 protein expression is observed in activated mouse liver myofibroblasts during hepatic fibrosis. Furthermore, Elf2 expression by activated liver myofibroblasts was confirmed, as Nt5e and Elf2 mRNA expression was both detected in both quiescent (2 days) and activated (28 days) primary mouse HSC cultured in vitro (Fig. 7a). Interestingly, culture-induced activation of primary mouse HSC led to an estimated 10-time decrease in Nt5e gene expression (quiescent, 101.6 ± 13.04 vs. activated, 9.8 ± 4.9, ***p < 0.001) and, in contrast, to an estimated two-time increase in Elf2 gene expression (quiescent, 101.5 ± 12.94 vs. activated, 207.1 ± 30.87, *p < 0.05) (Fig. 7a). We then tested the DNA motif requirements for Elf2 binding to the putative mouse Nt5e promoter sequence by performing electrophoretic mobility shift assays (using oligonucleotide probes mimicking the identified Elf2 promoter elements) in well-characterized immortalized JS1 and Col-GFP liver myofibroblasts that exhibit activated cell phenotype [25, 26]. Initially, expression of both Cd73 and Elf2 at the mRNA level was confirmed in both liver myofibroblast cell lines used (Fig. 7b). When nuclear extracts isolated from JS1 and Col-GFP cells were incubated with a digoxigenin-labeled wild-type EMSA probe containing a putative Elf2 binding site, multiple bands corresponding to formed DNA-nuclear protein complexes were observed (Fig. 7c, left panel). Next, the binding specificity was confirmed after addition of cold (unlabeled) wild-type probe in excess (125-fold molar), which considerably decreased signal detection. Interestingly, similar results were obtained for competitive binding experiments with cold probes harboring previously published Ets-related consensus sequences for Drosophila E74 gene promoter and murine blk gene promoter binding sites (not shown). These findings were further validated when incubation of JS1 and Col-GFP nuclear extracts with a mutated EMSA probe containing a disrupted putative Elf2 binding site did not results in the formation of DNA-nuclear protein complexes (Fig. 7c, right panel). These results demonstrate that mouse Cd73-expressing cells including liver myofibroblasts express regulatory transcription factors that can bind the identified Elf2 motif contained in the minimal mouse Nt5e promoter. Taken together, these data suggest that the activity of the putative minimal mouse Nt5e promoter is regulated by Elf2-like transcription factor(s).
Fig. 6.
Experimental liver fibrosis was induced by carbon tetrachloride (CCl4) intoxication for 3 weeks. In serial sections of livers from control (olive oil-injected) animals, normal expression of αSMA (blood vessels), Cd73 (parenchymal, sinusoidal areas), and Elf2 (non-parenchymal area) is depicted. In serial sections of livers from fibrotic (CCl4-intoxicated) animals, significant accumulation of αSMA protein is observed in lesion areas surrounding parenchymal cell nodules, along with Cd73 and Elf2 proteins. Scale bar 200 μm
Fig. 7.
a Relative mRNA expression of Nt5e and Elf2 genes is determined by quantitative real-time PCR analysis in quiescent and activated primary mouse HSC; mouse Gapdh was used as reference gene. b Expression of Nt5e and Elf2 mRNAs is determined by semi-quantitative PCR analysis in immortalized activated JS1 and Col-GFP mouse HSC, total mouse liver, and water (reaction control); mouse Gapdh was used as reference gene. c Electro-motility shift assays were performed using nuclear extracts obtained from mouse JS1 and Col-GFP cells, and digoxigenin-labeled probes corresponding to the minimal mouse Nt5e promoter containing the wild-type (Nt5e wt Elf2 probe, left panel) and mutated (Nt5e mut Elf2 probe, right panel) Elf2 motif (nucleotides −132 to −102) (see Table 1 for sequences). Competitive analysis was performed in the presence of a 125-fold molar excess of unlabeled competitors, as indicated. Arrowheads indicate formation of DNA-protein complexes (depicted image is representative of three independent experiments)
Discussion
In the present study, we report the cloning and characterization of the putative mouse Nt5e gene promoter. We have previously shown that Cd73 is expressed by activated liver myofibroblasts in both human and rat species and, now, show that it also applies in the mouse species using the classical CCl4 model of experimental liver fibrosis and both primary isolated and immortalized activated/myofibroblastic HSC. Using site-directed mutagenesis, we show that the sequence corresponding to the minimal mouse Nt5e gene promoter activity possesses a response element for Elf2-like transcription factor(s), acting as repressor upon binding. Fittingly, we further show that quiescent and activated primary isolated mouse HSC express both Nt5e and Elf2 gene products and their respective expression levels (Nt5e, down) and (Elf2, up) are inversely regulated during culture-induced activation. We also establish that nuclear extracts from Cd73-expressing activated liver JS1 and Col-GFP myofibroblasts contain Elf2-like transcription factor(s) that can bind to the minimal mouse Nt5e gene promoter. Taken together, our data suggest that Elf2-like transcription factor(s) mediate down-regulation of Nt5e gene expression in activated mouse HSC.
Our observation that Nt5e gene expression is down-regulated in mouse-activated liver myofibroblasts is directly in line with a previous study providing evidence that in vitro “myofibroblastic-to-quiescent” phenotype conversion of mouse-immortalized GRX myofibroblastic HSC induced by exogenous retinol treatment is accompanied with an increase of Nt5e gene expression [34]. Remarkably, Elf2 is a member of “E-twenty-six” (Ets)-related transcription factor family that all recognize a core DNA motif GGAA/T [35], akin to the GGAA element we have identified in the putative minimal mouse Nt5e gene promoter. Moreover, Ets-related transcription factors act as constitutive activators as well as repressors of genes involved in key cellular processes including proliferation, maturation, and differentiation [35, 36] that are taking place in liver myofibroblasts upon activation triggered by hepatic insult or injury [37]. Interestingly, examples of transcriptional regulation of “fibrogenic” genes by Ets-related transcription factors have been previously reported [38]. For instance, numerous studies have shown that the well-characterized Ets-related transcription factors are implicated in the regulation of matrix metalloproteinases [39], tissue inhibitors of metalloproteinases [40], tenascin [41], collagen [42], connective tissue growth factor [43] gene expression in activated skin-, lung-, and/or liver-derived stromal fibroblasts. While our results imply that Elf2-like transcription factor(s) can negatively regulate the putative mouse Nt5e gene promoter activity, it nevertheless remains challenging to predict the exact nature of DNA interaction and promoter-activity regulation occurring here. Importantly, gene expression regulation by Ets-related transcription factors is being rendered complex for several reasons. First, in vitro promoter studies performed have shown that multiple Ets-related transcription factors either positively or negatively promoting target gene expression can be expressed in the same cell type [30], and that promoter binding overlap exists between Ets-related transcription factors [44]. Additionally, in silico analysis of the full putative mouse Nt5e promoter reveals the presence of potential binding sites for Ets-related Elk1 and ERG transcription factors. Second, Ets-related transcription factors can be regulated by post-translational modifications such as phosphorylation [45] and glycosylation [46] that modulate their DNA-binding ability and function(s) [35]. For instance, a recent study has shown that transgenic mice overexpressing a mutant form of Ets2 transcription factor that cannot be site-specific phosphorylated are protected against experimental lung fibrosis when compared to wild-type animals [47]. Indeed, comparative protein sequence analysis indicates the existence of Elf2 splice variants that are lacking potential regulatory N-terminal phosphorylation sites [30]. Finally, the biological functions Ets-related transcription factors are usually dependent on the presence of another co-regulatory transcription factor [35, 36]. Overall, our results indicate that down-regulation of Nt5e gene expression in activated liver myofibroblasts is controlled by Elf2-like transcription factor(s). However, the possibility that additional mechanisms of regulation could be conferred by potential interacting partner transcription factor(s) or own Elf2-like transcription factor gene expression regulation in cell- and stage-dependent manner cannot be excluded. Importantly, in silico analysis of putative human and rat minimal promoter sequences did not reveal the existence of putative binding sites for Elf2 transcription factor or other Ets-related transcription factors (not shown).
We have also identified a potential enhancer region (−2069 to −1514 bp relative to mouse Cd73 ATG start codon). Luciferase assays show that the enhancer sequence can actually increase the activity of the putative minimal mouse Nt5e gene promoter, but cannot initiate transcription on its own. Moreover, in preliminary experiments in which we generated and used four consecutive truncations of the putative enhancer sequence for luciferase activity assays, we were not able to determine that a particular portion (out of the four analyzed) conferred the enhancing activity, as none of the constructs tested exhibited detectable levels of activity (not shown). Further investigation will be needed to identify what are the exact nature and identity of the transcription factors responsible for the described enhancer activity.
Lastly, our data showing down-regulation of mouse Nt5e gene expression in fibrotic liver and activated myofibroblasts are in agreement with previous reports describing an overall decrease in Nt5e gene expression is observed in experimental mouse models of chronic liver injury such as chronically feeding with cholestasis inducer 3,5-diethoxycarbonyl-1,4-dihydrocollidine compound or long-term exposure to oxidative stress inducer cerium trichloride [48, 49]. Additionally, human liver NT5E mRNA expression is decreased in biopsies from patients with established fibrosis secondary to hepatitis C infection or non-alcoholic fatty liver disease [48]. In contrast, up-regulation of mouse Nt5e gene expression has been reported in experimental models acute liver injury such as ethanol-induced steatosis and thioacetamide-induced acute liver failure [50, 51]. Moreover, observations made by our laboratory and others have shown up-regulation of rat Nt5e gene expression occurs in fibrotic livers and activated myofibroblasts, subjected to carbon tetrachloride intoxication, or exposed to low-dose ionizing radiation [15, 16, 52, 53]. Altogether, these findings may simply reflect differences in injury mechanisms associated with acute versus chronic liver diseases, as well as spatio-temporal regulation of Nt5e gene between species, indicating that mouse species is more suitable than rat species, to investigate the role of Cd73 biological activity in chronic liver diseases settings.
In conclusion, we establish that Cd73 is expressed by activated mouse liver myofibroblasts during hepatic fibrosis and show that down-regulation of Nt5e gene expression is partly mediated by Elf2-like Ets-related transcription factor(s) in these non-parenchymal cells. A better understanding of Nt5e gene expression and regulation may provide a new hindsight in the pathogenesis of liver fibrosis and lead to the development of novel anti-fibrotic therapies.
Acknowledgements
Funding was provided by Gilead Sciences Research Scholars Program in Liver Disease Award UAMS# 271G141616-01 to MF, NIH – NIDDK R56DK076735 to JAD, and NIH – NCRR/NIH –NCATS #UL1TR000039 - University of Arkansas for Medical Sciences Institutional Support.
Author contributions
MF designed, performed, and analyzed the experiments shown in Figs. 1, 2, 3, 4, 5, 6, conceived the study, and wrote the paper. EGL designed, performed, and analyzed the experiments shown in Figs. 1 and 6. JRG performed and provided technical assistance for experiments shown in Figs. 1 and 6. JAD conceived the study and edited the paper. All authors reviewed the results and approved the final version of the manuscript.
Compliance with ethical standards
Conflicts of interest
Michel Fausther declares that he has no conflict of interest.
Elise G. Lavoie declares that she has no conflict of interest.
Jessica R. Goree declares that she has no conflict of interest.
Jonathan A. Dranoff declares that he has no conflict of interest.
Ethical approval
All mouse experiments were performed in accordance with regulations approved by the University of Arkansas for Medical Sciences Institutional Animal Care and Use Committee.
References
- 1.Lee YA, Wallace MC, Friedman SL. Pathobiology of liver fibrosis: a translational success story. Gut. 2015;64:830–841. doi: 10.1136/gutjnl-2014-306842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hernandez-Gea V, Friedman SL. Pathogenesis of liver fibrosis. Annu Rev Pathol. 2011;6:425–456. doi: 10.1146/annurev-pathol-011110-130246. [DOI] [PubMed] [Google Scholar]
- 3.Novo E, Cannito S, Morello E, Paternostro C, Bocca C, Miglietta A, Parola M. Hepatic myofibroblasts and fibrogenic progression of chronic liver diseases. Histol Histopathol. 2015;30:1011–1032. doi: 10.14670/HH-11-623. [DOI] [PubMed] [Google Scholar]
- 4.Fausther M, Lavoie EG, Dranoff JA. Contribution of myofibroblasts of different origins to liver fibrosis. Curr Pathobiol Rep. 2013;1:225–230. doi: 10.1007/s40139-013-0020-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu J, Liu X, Koyama Y, Wang P, Lan T, Kim IG, Kim IH, Ma HY, Kisseleva T. The types of hepatic myofibroblasts contributing to liver fibrosis of different etiologies. Front Pharmacol. 2014;5:167. doi: 10.3389/fphar.2014.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferrari D, Gambari R, Idzko M, Muller T, Albanesi C, Pastore S, La Manna G, Robson SC, and Cronstein B (2015) Purinergic signaling in scarring. FASEB J [DOI] [PMC free article] [PubMed]
- 7.Wang H, Guan W, Yang W, Wang Q, Zhao H, Yang F, Lv X, Li J. Caffeine inhibits the activation of hepatic stellate cells induced by acetaldehyde via adenosine A2A receptor mediated by the cAMP/PKA/SRC/ERK1/2/P38 MAPK signal pathway. PLoS One. 2014;9:e92482. doi: 10.1371/journal.pone.0092482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Che J, Chan ES, Cronstein BN. Adenosine A2A receptor occupancy stimulates collagen expression by hepatic stellate cells via pathways involving protein kinase A, Src, and extracellular signal-regulated kinases 1/2 signaling cascade or p38 mitogen-activated protein kinase signaling pathway. Mol Pharmacol. 2007;72:1626–1636. doi: 10.1124/mol.107.038760. [DOI] [PubMed] [Google Scholar]
- 9.Yang P, Han Z, Chen P, Zhu L, Wang S, Hua Z, Zhang J. A contradictory role of A1 adenosine receptor in carbon tetrachloride- and bile duct ligation-induced liver fibrosis in mice. J Pharmacol Exp Ther. 2010;332:747–754. doi: 10.1124/jpet.109.162727. [DOI] [PubMed] [Google Scholar]
- 10.Chan ES, Montesinos MC, Fernandez P, Desai A, Delano DL, Yee H, Reiss AB, Pillinger MH, Chen JF, Schwarzschild MA, Friedman SL, Cronstein BN. Adenosine A(2A) receptors play a role in the pathogenesis of hepatic cirrhosis. Br J Pharmacol. 2006;148:1144–1155. doi: 10.1038/sj.bjp.0706812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsu SJ, Lee FY, Wang SS, Hsin IF, Lin TY, Huang HC, Chang CC, Chuang CL, Ho HL, Lin HC, Lee SD. Caffeine ameliorates hemodynamic derangements and portosystemic collaterals in cirrhotic rats. Hepatology. 2015;61:1672–1684. doi: 10.1002/hep.27679. [DOI] [PubMed] [Google Scholar]
- 12.Vecchio EA, White PJ and May LT (2017) Targeting Adenosine Receptors for the Treatment of Cardiac Fibrosis. Front Pharmacol 8:243. doi:10.3389/fphar.2017.00243 [DOI] [PMC free article] [PubMed]
- 13.Colgan SP, Eltzschig HK, Eckle T, Thompson LF. Physiological roles for ecto-5′-nucleotidase (CD73) Purinergic Signal. 2006;2:351–360. doi: 10.1007/s11302-005-5302-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fausther M, Lecka J, Soliman E, Kauffenstein G, Pelletier J, Sheung N, Dranoff JA, Sevigny J. Coexpression of ecto-5′-nucleotidase/CD73 with specific NTPDases differentially regulates adenosine formation in the rat liver. Am J Physiol Gastrointest Liver Physiol. 2012;302:G447–G459. doi: 10.1152/ajpgi.00165.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fausther M, Sheung N, Saiman Y, Bansal MB, Dranoff JA. Activated hepatic stellate cells upregulate transcription of ecto-5′-nucleotidase/CD73 via specific SP1 and SMAD promoter elements. Am J Physiol Gastrointest Liver Physiol. 2012;303:G904–G914. doi: 10.1152/ajpgi.00015.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ji J, Yu F, Ji Q, Li Z, Wang K, Zhang J, Lu J, Chen L, E Q, Zeng Y, Ji Y. Comparative proteomic analysis of rat hepatic stellate cell activation: a comprehensive view and suppressed immune response. Hepatology (Baltimore, Md) 2012;56:332–349. doi: 10.1002/hep.25650. [DOI] [PubMed] [Google Scholar]
- 17.Berardis S, Lombard C, Evraerts J, El Taghdouini A, Rosseels V, Sancho-Bru P, Lozano JJ, van Grunsven L, Sokal E, Najimi M. Gene expression profiling and secretome analysis differentiate adult-derived human liver stem/progenitor cells and human hepatic stellate cells. PLoS One. 2014;9:e86137. doi: 10.1371/journal.pone.0086137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koyama Y, Wang P, Liang S, Iwaisako K, Liu X, Xu J, Zhang M, Sun M, Cong M, Karin D, Taura K, Benner C, Heinz S, Bera T, Brenner DA, Kisseleva T. Mesothelin/mucin 16 signaling in activated portal fibroblasts regulates cholestatic liver fibrosis. J Clin Invest. 2017;127:1254–1270. doi: 10.1172/JCI88845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Synnestvedt K, Furuta GT, Comerford KM, Louis N, Karhausen J, Eltzschig HK, Hansen KR, Thompson LF, Colgan SP. Ecto-5′-nucleotidase (CD73) regulation by hypoxia-inducible factor-1 mediates permeability changes in intestinal epithelia. J Clin Invest. 2002;110:993–1002. doi: 10.1172/JCI0215337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chalmin F, Mignot G, Bruchard M, Chevriaux A, Vegran F, Hichami A, Ladoire S, Derangere V, Vincent J, Masson D, Robson SC, Eberl G, Pallandre JR, Borg C, Ryffel B, Apetoh L, Rebe C, Ghiringhelli F. Stat3 and Gfi-1 transcription factors control Th17 cell immunosuppressive activity via the regulation of ectonucleotidase expression. Immunity. 2012;36:362–373. doi: 10.1016/j.immuni.2011.12.019. [DOI] [PubMed] [Google Scholar]
- 21.Regateiro FS, Cobbold SP, Waldmann H. CD73 and adenosine generation in the creation of regulatory microenvironments. Clin Exp Immunol. 2013;171:1–7. doi: 10.1111/j.1365-2249.2012.04623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng Z, Fernandez P, Wilder T, Yee H, Chiriboga L, Chan ES, Cronstein BN. Ecto-5′-nucleotidase (CD73)-mediated extracellular adenosine production plays a critical role in hepatic fibrosis. FASEB J. 2008;22:2263–2272. doi: 10.1096/fj.07-100685. [DOI] [PubMed] [Google Scholar]
- 23.Kruglov EA, Jain D, Dranoff JA. Isolation of primary rat liver fibroblasts. J Investig Med. 2002;50:179–184. doi: 10.2310/6650.2002.33431. [DOI] [PubMed] [Google Scholar]
- 24.Jainchill JL, Aaronson SA, Todaro GJ. Murine sarcoma and leukemia viruses: assay using clonal lines of contact-inhibited mouse cells. J Virol. 1969;4:549–553. doi: 10.1128/jvi.4.5.549-553.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo J, Loke J, Zheng F, Hong F, Yea S, Fukata M, Tarocchi M, Abar OT, Huang H, Sninsky JJ, Friedman SL. Functional linkage of cirrhosis-predictive single nucleotide polymorphisms of Toll-like receptor 4 to hepatic stellate cell responses. Hepatology. 2009;49:960–968. doi: 10.1002/hep.22697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meurer SK, Alsamman M, Sahin H, Wasmuth HE, Kisseleva T, Brenner DA, Trautwein C, Weiskirchen R, Scholten D. Overexpression of endoglin modulates TGF-beta1-signalling pathways in a novel immortalized mouse hepatic stellate cell line. PLoS One. 2013;8:e56116. doi: 10.1371/journal.pone.0056116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weiskirchen R, Gressner AM. Isolation and culture of hepatic stellate cells. Methods Mol Med. 2005;117:99–113. doi: 10.1385/1-59259-940-0:099. [DOI] [PubMed] [Google Scholar]
- 28.Dranoff JA, Ogawa M, Kruglov EA, Gaca MD, Sevigny J, Robson SC, Wells RG. Expression of P2Y nucleotide receptors and ectonucleotidases in quiescent and activated rat hepatic stellate cells. Am J Physiol Gastrointest Liver Physiol. 2004;287:G417–G424. doi: 10.1152/ajpgi.00294.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Oettgen P, Akbarali Y, Boltax J, Best J, Kunsch C, Libermann TA. Characterization of NERF, a novel transcription factor related to the Ets factor ELF-1. Mol Cell Biol. 1996;16:5091–5106. doi: 10.1128/MCB.16.9.5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chrisman HR, Tindall DJ. Identification and characterization of a consensus DNA binding element for the zinc finger transcription factor TIEG/EGRalpha. DNA Cell Biol. 2003;22:187–199. doi: 10.1089/104454903321655819. [DOI] [PubMed] [Google Scholar]
- 32.Karsenty G, de Crombrugghe B. Conservation of binding sites for regulatory factors in the coordinately expressed alpha 1 (I) and alpha 2 (I) collagen promoters. Biochem Biophys Res Commun. 1991;177:538–544. doi: 10.1016/0006-291X(91)92017-E. [DOI] [PubMed] [Google Scholar]
- 33.McPherson LA, Weigel RJ. AP2alpha and AP2gamma: a comparison of binding site specificity and trans-activation of the estrogen receptor promoter and single site promoter constructs. Nucleic Acids Res. 1999;27:4040–4049. doi: 10.1093/nar/27.20.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andrade CM, Roesch GC, Wink MR, Guimaraes EL, Souza LF, Jardim FR, Guaragna RM, Bernard EA, Margis R, Borojevic R, Battastini AM, Guma FC. Activity and expression of ecto-5′-nucleotidase/CD73 are increased during phenotype conversion of a hepatic stellate cell line. Life Sci. 2008;82:21–29. doi: 10.1016/j.lfs.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 35.Sharrocks AD. The ETS-domain transcription factor family. Nat Rev Mol Cell Biol. 2001;2:827–837. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- 36.Oikawa T, Yamada T. Molecular biology of the Ets family of transcription factors. Gene. 2003;303:11–34. doi: 10.1016/S0378-1119(02)01156-3. [DOI] [PubMed] [Google Scholar]
- 37.Friedman SL. Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol Rev. 2008;88:125–172. doi: 10.1152/physrev.00013.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hahne JC, Okuducu AF, Fuchs T, Florin A, Wernert N. Identification of ETS-1 target genes in human fibroblasts. Int J Oncol. 2011;38:1645–1652. doi: 10.3892/ijo.2011.981. [DOI] [PubMed] [Google Scholar]
- 39.Ozaki I, Zhao G, Mizuta T, Ogawa Y, Hara T, Kajihara S, Hisatomi A, Sakai T, Yamamoto K. Hepatocyte growth factor induces collagenase (matrix metalloproteinase-1) via the transcription factor Ets-1 in human hepatic stellate cell line. J Hepatol. 2002;36:169–178. doi: 10.1016/S0168-8278(01)00245-8. [DOI] [PubMed] [Google Scholar]
- 40.Hahne JC, Fuchs T, El Mustapha H, Okuducu AF, Bories JC, Wernert N. Expression pattern of matrix metalloproteinase and TIMP genes in fibroblasts derived from Ets-1 knock-out mice compared to wild-type mouse fibroblasts. Int J Mol Med. 2006;18:153–159. [PubMed] [Google Scholar]
- 41.Shirasaki F, Makhluf HA, LeRoy C, Watson DK, Trojanowska M. Ets transcription factors cooperate with Sp1 to activate the human tenascin-C promoter. Oncogene. 1999;18:7755–7764. doi: 10.1038/sj.onc.1203360. [DOI] [PubMed] [Google Scholar]
- 42.Czuwara-Ladykowska J, Shirasaki F, Jackers P, Watson DK, Trojanowska M. Fli-1 inhibits collagen type I production in dermal fibroblasts via an Sp1-dependent pathway. J Biol Chem. 2001;276:20839–20848. doi: 10.1074/jbc.M010133200. [DOI] [PubMed] [Google Scholar]
- 43.Leask A, Chen S, Pala D, Brigstock DR. Regulation of CCN2 mRNA expression and promoter activity in activated hepatic stellate cells. J Cell Commun Signal. 2008;2:49–56. doi: 10.1007/s12079-008-0029-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hollenhorst PC, Shah AA, Hopkins C, Graves BJ. Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes Dev. 2007;21:1882–1894. doi: 10.1101/gad.1561707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Selvaraj N, Kedage V, Hollenhorst PC. Comparison of MAPK specificity across the ETS transcription factor family identifies a high-affinity ERK interaction required for ERG function in prostate cells. Cell Commun Signal. 2015;13:12. doi: 10.1186/s12964-015-0089-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tootle TL, Rebay I. Post-translational modifications influence transcription factor activity: a view from the ETS superfamily. BioEssays. 2005;27:285–298. doi: 10.1002/bies.20198. [DOI] [PubMed] [Google Scholar]
- 47.Baran CP, Fischer SN, Nuovo GJ, Kabbout MN, Hitchcock CL, Bringardner BD, McMaken S, Newland CA, Cantemir-Stone CZ, Phillips GS, Ostrowski MC, Marsh CB. Transcription factor ets-2 plays an important role in the pathogenesis of pulmonary fibrosis. Am J Respir Cell Mol Biol. 2011;45:999–1006. doi: 10.1165/rcmb.2010-0490OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Snider NT, Griggs NW, Singla A, Moons DS, Weerasinghe SV, Lok AS, Ruan C, Burant CF, Conjeevaram HS, Omary MB. CD73 (ecto-5′-nucleotidase) hepatocyte levels differ across mouse strains and contribute to mallory-denk body formation. Hepatology. 2013;58:1790–1800. doi: 10.1002/hep.26525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheng J, Fei M, Fei M, Sang X, Sang X, Cheng Z, Gui S, Zhao X, Sheng L, Sun Q, Hu R, Wang L, Hong F. Gene expression profile in chronic mouse liver injury caused by long-term exposure to CeCl3. Environ Toxicol. 2014;29:837–846. doi: 10.1002/tox.21826. [DOI] [PubMed] [Google Scholar]
- 50.Peng Z, Borea PA, Varani K, Wilder T, Yee H, Chiriboga L, Blackburn MR, Azzena G, Resta G, Cronstein BN. Adenosine signaling contributes to ethanol-induced fatty liver in mice. J Clin Invest. 2009;119:582–594. doi: 10.1172/JCI37409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hart ML, Grenz A, Gorzolla IC, Schittenhelm J, Dalton JH, Eltzschig HK. Hypoxia-inducible factor-1alpha-dependent protection from intestinal ischemia/reperfusion injury involves ecto-5′-nucleotidase (CD73) and the A2B adenosine receptor. J Immunol. 2011;186:4367–4374. doi: 10.4049/jimmunol.0903617. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 52.Kristensen DB, Kawada N, Imamura K, Miyamoto Y, Tateno C, Seki S, Kuroki T, Yoshizato K. Proteome analysis of rat hepatic stellate cells. Hepatology (Baltimore, Md) 2000;32:268–277. doi: 10.1053/jhep.2000.9322. [DOI] [PubMed] [Google Scholar]
- 53.Schmid TC, Loffing J, Le Hir M, Kaissling B. Distribution of ecto-5′-nucleotidase in the rat liver: effect of anaemia. Histochemistry. 1994;101:439–447. doi: 10.1007/BF00269494. [DOI] [PubMed] [Google Scholar]