FIGURE 1.
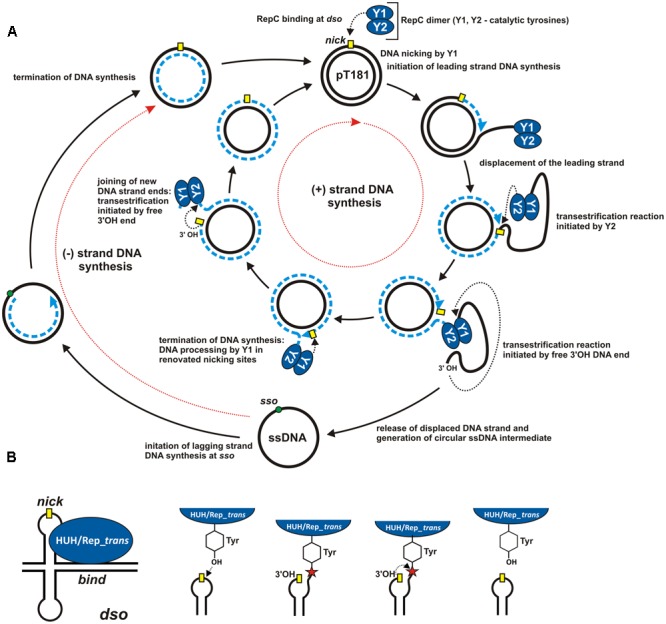
Mechanism of rolling-circle replication. (A) A model for RCR based on studies of plasmid pT181. Separated cycles of leading (+) and lagging (–) DNA strand synthesis (initiated at the dso and sso origins, respectively) are marked by red dotted arrows. The individual steps in the synthesis are described in the Figure and details are given in the text. The RCR initiation protein (RepC) initiates (+) strand replication at the nick site (marked by a yellow rectangle). Dashed blue lines indicate the nascent DNA strand and blue arrows show the direction of synthesis. Black dotted arrows indicate nucleophilic attack of the catalytic tyrosine residue on the phosphodiester bond in the DNA or of the free 3′-OH DNA end on the phosphotyrosine bond between the enzyme and the DNA. (B) ssDNA breakage and joining reactions catalyzed by HUH and Rep_trans endonucleases at the dso. The nick site exposed in a hairpin-like structure is shown by a yellow rectangle. The red star indicates the phosphotyrosine bond between the enzyme and the 5′-end of the DNA (see text for details). The reaction catalyzed by HUH/Rep_trans is reversible since the released 3′-OH DNA end could acts as a nucleophile on the phosphotyrosine bond (adopted from Chandler et al., 2013; Pastrana et al., 2016).
