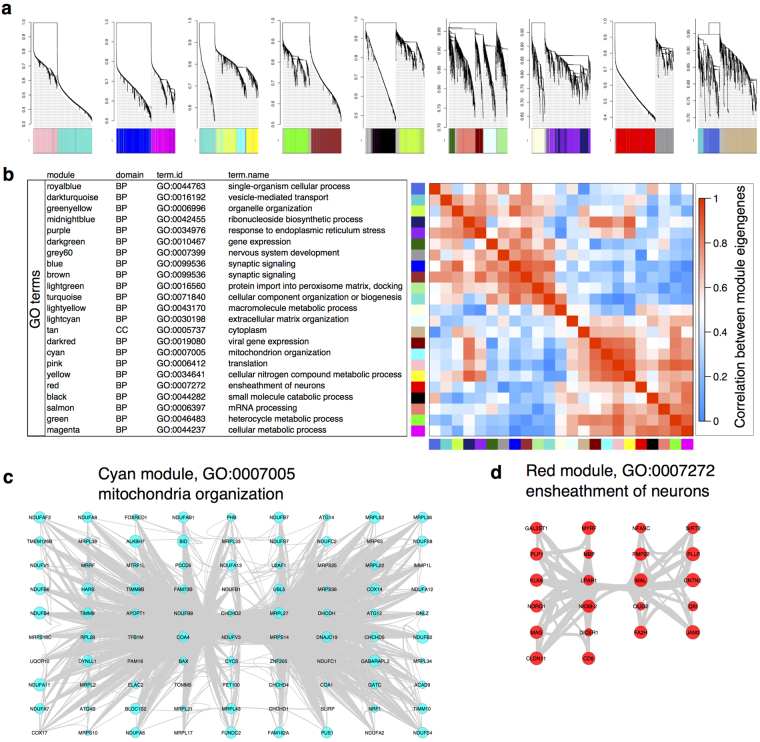
Figure 1.
WGCNA derived modules of gene expression. (a) An overall view of the network of gene expression. Height on the y-axes represents the dissimilarity of detected transcripts from the overall topology of gene expression. Colors below each line show the modules to which each transcript was assigned. (b) The eigengenes for each module are enriched for specific biological processes. For each module, we performed gene ontology (GO) enrichment, listing the term.id and name for each module (see also supplementary file 4 for p values). The module eigengenes showed correlation with each other, as indicated in the heatmap on the right. (c,d) Examples of modules enriched for biological terms. We used cytoscape to visualize organization of genes assigned to the GO category mitochondria organization in the cyan module (c) or the GO category ‘ensheathment of neurons’ in the red module (d). Circles are sized by the within module connectivity (Kwithin).