Figure 1.
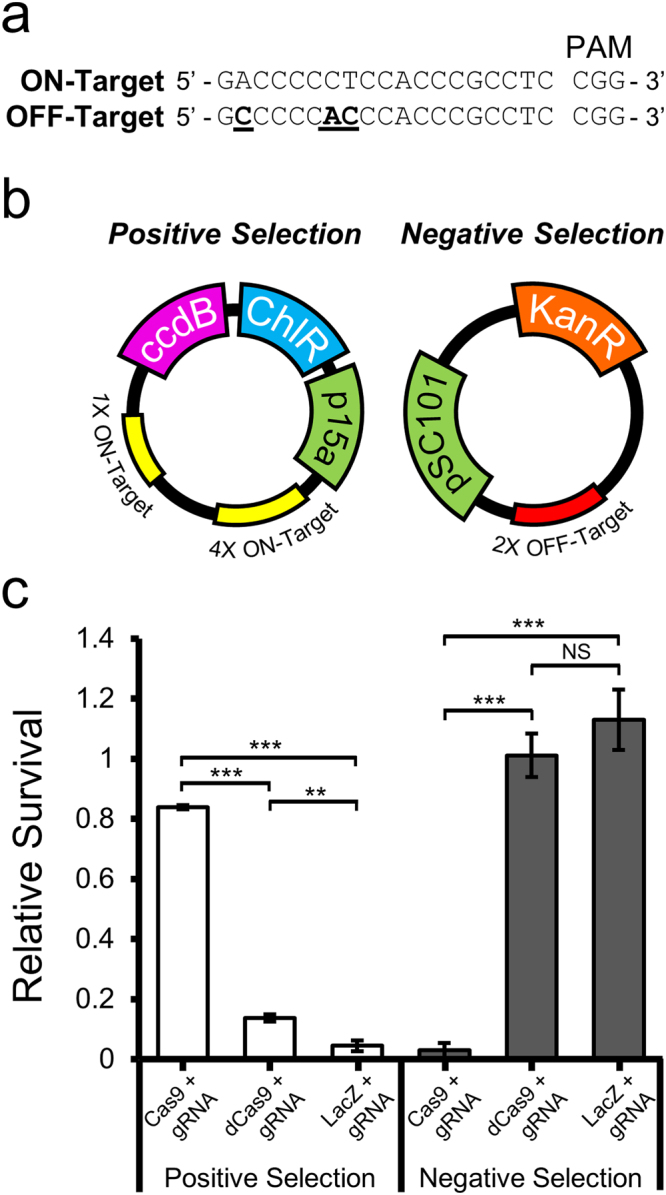
Positive and Negative Selection Systems. (a) A target sequence with known off-target activity was chosen to insert into the positive selection plasmid (ON-target). One of the most highly active off-target sequences was inserted into the negative selection plasmid (OFF-target). Mismatched bases are underlined and highlighted in bold. (b) Cartoon representation of positive and negative selction plasmids. The positive selection plasmid expresses ccdB under control of the Ptac promoter, carries a chloramphenicol resistance gene, and controls copy number with the p15a origin of replication. Yellow boxes indicate sites where four or a single copy of the ON-target sequence have been inserted into the plasmid. The low-copy, negative selection plasmid includes a kanamycin resistance gene and controls its copy number with the pSC101 origin of replication. The red box indicates the insertion of two copies of the OFF-target sequence. (c) Selection plasmids were cotransformed with a plasmid expressing the on-target gRNA and either active Cas9, nuclease dead Cas9 (dCas9) or LacZ. Comparison of transformations plated on selective media to control plates were used to assess relative survival. The Cas9 plasmid showed significantly greater rescue on selection media than dCas9 or LacZ (n = 3, error bars indicate S.E.M., one-way ANOVA F2,6 = 1181, P = 1.63 × 10−8, followed by post-hoc Tukey HSD analysis). Cas9 expressing plasmid showed an inverse response on selection media (n = 3, error bars indicate S.E.M., one-way ANOVA F2,6 = 68.63, P = 7.35 × 10−5, followed by post-hoc Tukey HSD analysis, P = * < 0.05, ** < 0.01, *** < 0.001, NS = not significant).
