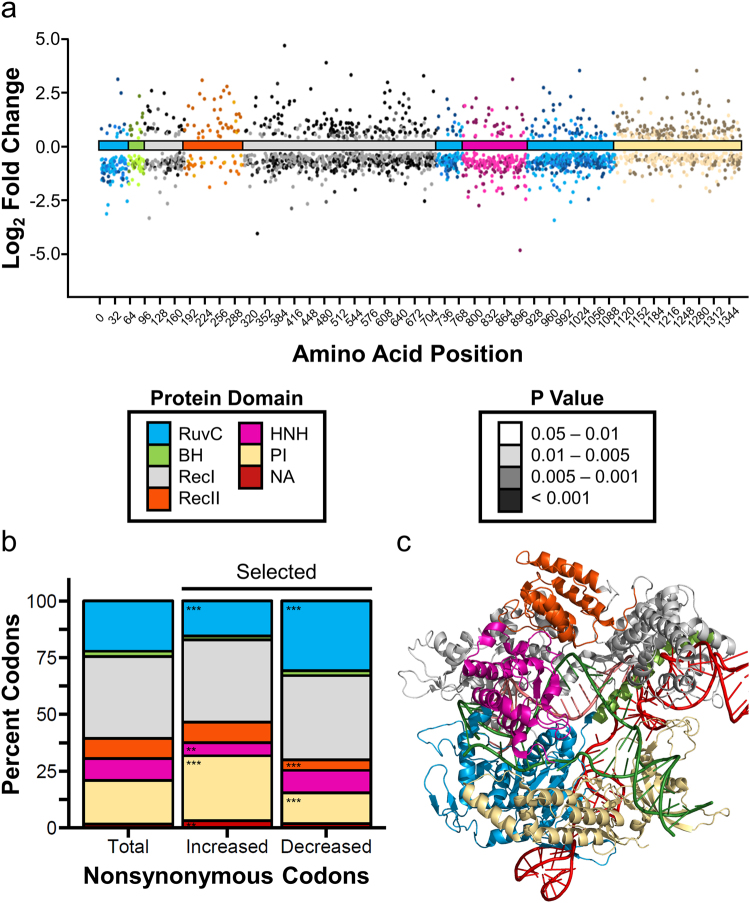
Figure 5.
Domains have varied responses to mutation. (a) The log2 transformed fold change in mutation frequency following positive selection is plotted across the length of SpCas9. Each point represents a unique nonsynonymous mutation that was significantly enriched or depleted following selection. Points are colored by protein domain. The shade of the color indicates P value as calculated by Fisher’s exact test adjusted using the Benjamini-Hochberg procedure. (b) Nonsynonymous codons that resulted from one nucleotide change were counted for each structural domain and represented as a percent of all possible nonsynonymous codons within the open reading frame of the protein (Total). Similarly, nonsynonymous codons which were significantly enriched (Increased) or depleted (Decreased) following selection were counted and represented as a percent of all significantly altered codons of each respective class. Chi-squared analysis was performed on the frequencies of significantly altered codons to determine which domains were more or less tolerant of mutations (P = ** < 0.01, *** < 0.001). (c) Cartoon structure of SpCas9 bound to gRNA and dsDNA based on PDB: 5f9r (ref.27). Domains are colored as in (a) and (b) for reference.