Figure 4.
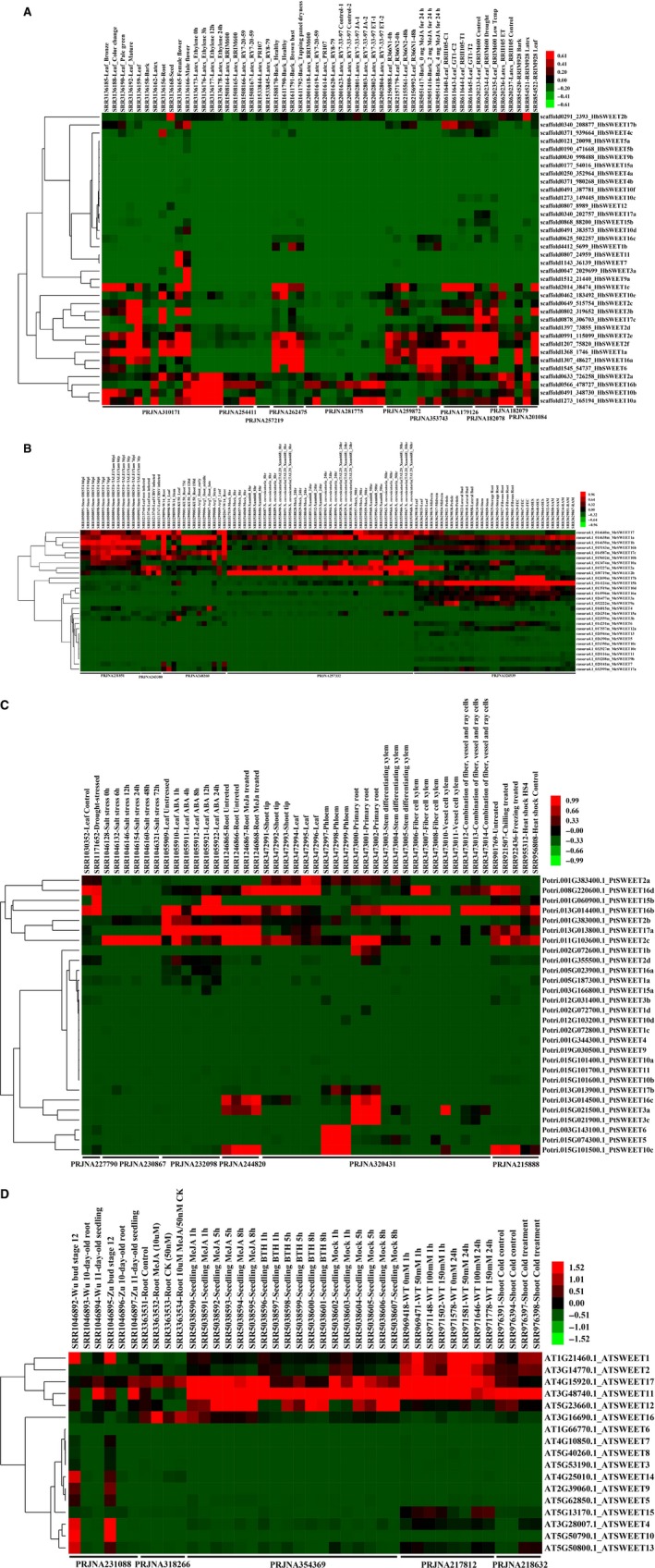
Expression analyses of the SWEET genes based on Solexa sequencing. (A), Hierarchical clustering and differential expression analysis of the HbSWEET genes in seven tissues (leaf, bark, latex, root, seed, female flower, and male flower), at four developmental stages of leaves (bronze, color change, pale‐green and mature), during ethephon treatment (0 h, 3 h, 12 h, and 24 h, PRJNA310171), latex (RRIM600 and RY7‐20‐59, PRJNA254411; clones PR107 and RY879, PRJNA257219), brown bast and tapping panel dryness (PRJNA262475), ET and JA treatment (PRJNA281775), Clone FX 3864 response to GCL012 (PRJNA259872), MeJA (PRJNA353743), Corynespora cassiicola tolerance (PRJNA179126), abiotic stress (drought, low temperature, PRJNA182078 and ethephon treatment, PRJNA182079), and tissues (leaf, bark, and latex, PRJNA201084); (B), Hierarchical clustering and differential expression analysis of the MeSWEET genes in different tissues (root, leaf, stem, PRJNA248260), infected by pathogenic Xanthomonas (PRJNA231851), CBSV virus (PRJNA243380), tissues (PRJNA324539), and bacterial blight pathogen (PRJNA257332); (C), Hierarchical clustering and differential expression analysis of the PtSWEET genes under ABA stimulation (0 h, 1 h, 4 h, 8 h, 12 h, and 24 h, PRJNA232098), methyl jasmonate stimulation (PRJNA244820), chilling, freezing, and heat shock (PRJNA207974, PRJNA215888), salinity stress (0 h, 6 h, 12 h, 24 h, and 72 h, PRJNA230867), tissues (PRJNA320431), drought stress (PRJNA227790); (D), Hierarchical clustering and differential expression analysis of the AtSWEET genes in different tissues (floral bud, root, seeding, PRJNA231088), MeJA or BTH (PRJNA354369), MeJA and CK (PRJNA318266), cold stress (PRJNA218632), salt stress (0 mm, 50 mm, 100 mm, 150 mm, PRJNA217812).
