Abstract
A highly effective antigen construct for presenting conserved antigen domains is essential to the development of a universal influenza vaccine. We have developed a novel dual-domain nanoparticle fusion protein (DDNFP) which allows independent presentation of two conserved domains. The conserved domains used were from two separate viral surface proteins, M2e of M2 and fusion peptide (FP) or long alpha helix (CD) of HA2. The carrier is a novel nanoparticle protein - the dodecameric DNA binding protein from starved cells (Dps) of bacteria or archaea. Dps was found to be uniquely capable of simultaneous fusion and surface presentation at both N- and C-termini while retaining the ability to form nanoparticles. Thus, DDNFPs with M2e and FP or CD fused at N- and C- termini of Dps from E. coli (EcDps) or other bacteria were first constructed based on the H1 subtype sequences along with corresponding single-domain nanoparticle fusion proteins (SDNFPs). They were expressed at high levels in bacteria and found to form nanoparticles of the expected size (~9 nm). They were stable against treatment at high temperatures. The DDNFPs (M2e-EcDps-FP and M2e-EcDps-CD) induced strong antibody responses against individual antigen domains and provided full protection against lethal challenge with PR8 virus (H1N1). Importantly, the protection by DDNFPs was synergistically enhanced as compared to SDNFPs. The M2e-EcDps-CD provided an even stronger protection than M2e-EcDps-FP and therefore appeared to be the superior construct. Together, with novel domain combination, enhanced protection and ease of production, this M2e/CD DDNFP could potentially be a highly effective antigen construct for the universal influenza vaccine.
Keywords: Influenza, vaccine, conserved domains, dual-domain, nanoparticle
Introduction
Influenza viruses undergo constant genetic and antigenic changes. There are 18 influenza A subtypes which fall into five clades within two broad phylogenetic groups [1]. Current trivalent and quadrivalent inactivated vaccines (TIV and QIV; H1, H3, and one or two B strains) for seasonal influenza are strain-specific and not suited for controlling pandemic or variant epidemic viruses. Thus, a universal influenza vaccine which is effective against divergent influenza viruses is greatly needed to provide better control of seasonal epidemics and an effective counter measure against pandemics. The basic strategy for developing such a vaccine is to target the conserved domains so that immune responses generated against them can be cross-protective against divergent influenza viruses [2–3]. The conserved domains such as M2e and those from HA2 have been major targets for universal influenza vaccine candidates [1][3]. In addition, the HA stem region recognized by broadly neutralizing antibodies has also been targeted with headless HA or whole stem constructs [4–6].
M2e is the 24-aa extracellular domain of the minor envelope protein M2 which is highly conserved among all influenza A viruses, especially within the first nine amino acids [7–8]. M2 has a low copy number per virion, but is present abundantly on infected cells [9]. It is one of the most widely studied targets for the universal influenza vaccine and has been shown to provide strong protection through antibody-dependent cell-mediated cytotoxicity (ADCC) [1,8]. The HA2 makes up the bulk of HA stem and consists of distinct domains including FP, A, B, C, D, E, F, G and H [10]. It is highly conserved within each or closely related subtypes, but varies between subtypes from different phylogenetic groups [11–12]. FP consists of the first 38 aa of HA2 and is conserved among both influenza A and B viruses with substitution found only at two positions (2 and 12) among its first 14 amino acids (GLFGAIAGFIEGGW) [7,13]. The CD domain, also referred as the long alpha helix, consists of aa 76 – 130 of HA2 which forms the central part of the HA stem. Both have been shown to induce the protective immune responses [14–17].
Various approaches have been used to target these conserved domains [3,18]. Different carriers including nanoparticles or virus-like particles have been evaluated, including Hbc [19], papaya mosaic virus [20], influenza virus NP protein [21], tuftsin [22], flagellin [23], bacterial phage [24], adenovirus vector [25], and influenza virus-like particle [26]. However, these efforts have been primarily based on the single-domain approach.
We selected the DNA binding protein from starved cells (Dps) as the carrier protein to generate an antigen construct which can combine two separate domains for enhanced protection. Dps is a nanoparticle protein (~9 nm) present exclusively in bacteria or archaea [27–28]. Each Dps particle consists of 12 identical subunits (~ 20 kDa each). Through evaluation of various influenza virus domains and Dps from different bacteria, we found that Dps is uniquely capable of simultaneous and separate fusion and surface presentation of antigen domains at both N- and C-termini. The resulting DDNFP incorporating M2e and a HA2 domain (FP or CD) were successfully expressed at high levels in E. coli. Importantly, the DDNFP generated balanced immune responses against individual domains and provided synergistically enhanced protection against the lethal challenge. It could therefore be a highly effective antigen construct for development of the universal influenza vaccine.
Materials and methods
Cloning, expression, and purification of Dps fusion proteins
The M2e, FP and CD domains were fused individually or in combination to N- and/or C-termini of Dps as SDNFP or DDNFP (Fig. 1). The amino acid sequences of the domains (M2e, FP, and CD) used are listed in Supplementary Table 1. The M2e is the consensus sequence of human influenza A viruses. FP and CD sequences were the consensus sequences of the H1 subtype. Two Dps carriers from E. coli (EcDps) and the hyperthermopile S. solfataricus (SsDps) were evaluated (Supplementary Table 2). The fusion protein genes were generated by DNA synthesis (DNA 2.0, Menlo Park, CA and Genscript, Piscataway, NJ) or PCR using appropriate primers for linking the domain to either N- or C-terminus of the Dps carrier. Proteins were expressed at either 37°C or room temperature (RT) in E. coli (BL21) using pET 11 or pJexpress (DNA 2.0, Menlo Park, CA) vector following induction with 1 mM IPTG. Bacterial cells were lysed by sonication in phosphate buffered saline (PBS). Purification was performed by ion exchange (Q-Sepharose) followed by the size exclusion chromatography (SEC, Sephacryl S300). Purified fusion proteins reached a purity of at least 90% by SDS-PAGE and densitometry analysis of the fusion protein and any host cell protein bands. Protein concentrations were determined by BCA assay.
Fig. 1.
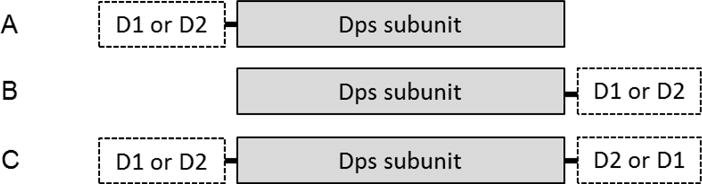
A schematic representation of Dps fusion proteins with influenza virus antigen domains. A and B, single-domain fusion proteins; C, dual-domain fusion protein. D1 and D2 represents two different domains which can be M2e, FP or CD.
Transmission electron microscopy
Transmission electron microscopy (TEM) was performed using the sodium phosphotungstate negative stain to examine the morphology of Dps and SDNFP and DDNFP at the Imaging Center of College of Veterinary Medicine and Biomedical Sciences, Texas A&M University.
Animals and challenge experiments
All animal studies were conducted with the approval of the Institutional Animal Use and Care Committee at Texas A&M University. The 6–8 week old Balb/c mice were obtained from Envigo (formerly Harlan Laboratories Inc). Groups of mice (n=5) were immunized by intramuscular injection with fusion proteins at 10 μg/mouse in combination with Sigma adjuvant system (SAS) (Sigma Chemical Co., St. Louis, IL; formerly Ribi adjuvant) in 50 μl, twice four weeks apart. The SAS, a squalene oil-in-water emulsion containing MPL (Monophosphoryl lipid A) and trehalose dicorynomycolate, was used by mixing with antigen at 1:1 ratio per the instruction provided by the manufacturer. It was more effective than Alhydrogel (Invivogen, San Diego, CA; 0.5% (w/v) or 250 μg/mouse) for enhancing antibody responses as shown in a comparison experiment with a DDNFP (M2e/FP) (Supplementary Fig. 3). Serum samples were collected every two weeks by submandibular bleeding technique till the end of the experiment. Challenge was performed intranasally with the PR8 virus (A/Puerto Rico/8/34, H1N1) at a lethal dose of 5 or 10 LD50 in 30 μl. The PR8 virus was propagated in MDCK cells and diluted to appropriate dose (LD50) with PBS. Mice were briefly anesthetized with isoflurane for nasal administration.
ELISA and neutralization test (NT)
ELISA was performed with 96-well plates (Maxisorp, Nunc). The plates were coated with synthetic peptides (2 μg/ml), inactivated whole virus (5 μg/ml, A/New Caledonia/20/1999, H1N1), or recombinant HA (1 μg/ml, A/New Caledonia/20/1999, H1N1; BEI Resources) in 0.1 M carbonate buffer (pH 9.6) at 4°C overnight. The inactivated whole virus antigen was prepared by purification from infected MDCK cells and inactivation with formaldehyde [29,30]. The synthetic peptides were M2e (24 aa), FP (38 aa), and C (29 aa) made by Genscript (Piscataway Township, NJ) or Peptide 2.0 (Chantilly, VA). The C peptide was used to measure the responses against CD. PBS-T buffer (20 mM phosphate, 150 mM NaCl, pH 7.4; 0.025% Tween 20) containing 3% BSA was used for blocking and sample dilution. The plates were blocked at RT for 2 hrs. Serum samples were serially 2-fold diluted and incubated at RT for 2 hrs. After washing, plates were incubated with anti-mouse IgG alkaline phosphatase conjugate (Sigma Chemical Co, St. Luis) at RT for 1 hr, which was followed by washing and incubation with PNPP substrate (Thermo Scientific Pierce) for 30 min. The OD was measured at 405 nm. The antibody titer was determined as the highest dilution with an OD value 2-fold above the background.
NT was performed with MDCK cells in 96-well plates as described in the WHO manual on animal influenza diagnosis and surveillance [31]. Briefly, pooled serum samples from each group were inactivated at 56°C for 30 min and serially diluted by 2-fold before mixing with 100 TCID50 of PR8 virus in duplicate. After incubation at 37°C for 1 hr, the mixtures were transferred to MDCK cells and incubated at 37°C for 1 hr. The plates were then washed twice with PBS after removing the mixtures, and fresh serum free media containing 2 μg/ml trypsin was added. The plates were incubated at 37°C for 72 hrs and then fixed in formalin and stained by crystal violet. The neutralization titer is the highest serum dilution with the intact cell monolayer.
Immunoblot
Antigen proteins were separated by SDS-PAGE and blotted onto Immobilon P polyvinylidene difluoride membranes (Millipore). They were mixed at 1:1 ratio with sample buffer (Bio-Rad) containing 2% SDS and 5% 2-mercaptoethanol and then kept in boiling water bath for 3 min prior to gel electrophoresis. Blotted membranes were first blocked at RT in PBS-T buffer containing 3% BSA for 2 hrs. They were then probed at RT for 2 hrs with the pooled serum sample diluted at 1:2,000 in the same buffer with BSA. After washing with PBS-T, they were incubated for 1 hr with the alkaline phosphatase-conjugated anti-mouse IgG Fc (Sigma Chemical Co., St. Luis). Membranes were developed with substrate BCIP (5-bromo-4-chloro-3-indolylphosphate)–NBT (nitroblue tetrazolium) (Sigma Chemical Co.) for 30 min. The densitometry was performed using NIH ImageJ (http://rsbweb.nih.gov/ij/) for comparing the optical densities of the positive bands on the same blot.
Statistics
Statistical analyses were performed using Microsoft Excel. The geometric mean titer (GMT) and the standard deviation were determined for each group. Means were compared using the student t test. The survival results were analyzed by the log-rank test [32]. A p value ≤0.05 is considered significant.
Results
Generation and Characterization of Dps fusion proteins with M2e and HA2 domains
A series of Dps fusion proteins with M2e, FP and/or CD domains at either or both of N-and C-termini were generated and evaluated (Fig. 1). Most of them were expressed at high levels as soluble proteins, especially at RT. The fusion proteins consistently exhibited an increased MW by SDS-PAGE that correlated with the addition of individual domains by fusion. A series of fusion proteins with EcDps are shown in Fig. 2A. A similar set of fusion proteins with SsDps were also generated (data not shown). It was noted that some fusion proteins with different domains, e.g., EcDps-FP and M2e-EcDps-FP, did not exhibit an apparent or proportional difference in migration or Mw as expected based on the domain number or length by SDS-PAGE (Fig. 2A). This might be due to a slight difference during protein denaturation or interaction with SDS as has been observed with other proteins [33].
Fig. 2.
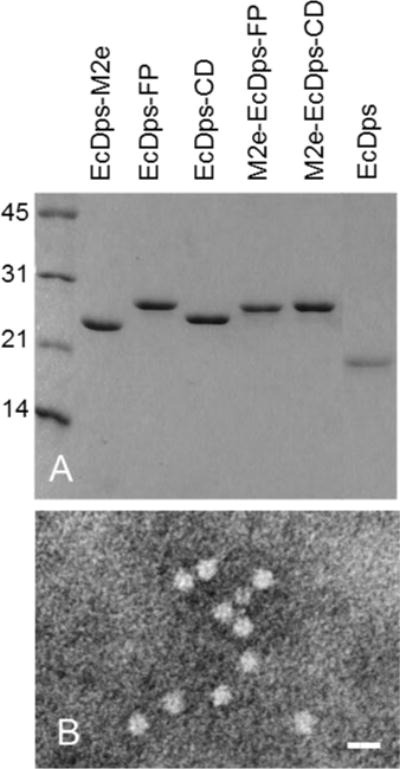
Analysis of purified fusion proteins by SDS-PAGE and electron microscopy (EM). A. SDS-PAGE; B, Electron micrograph of the DDNFP (M2e-EcDps-CD). Bar=20 nm.
All selected fusion proteins, including dual-domain fusion proteins, appeared as nanoparticles of expected size (~ 9 nm) as shown by TEM (Fig. 2B) and elution positions by SEC which were same as or faster than the native Dps. This indicated that Dps fusion proteins retained the ability to form nanoparticles with addition of antigen domains at either or both of N-and C-termini. They are therefore single- or dual-domain nanoparticle fusion proteins (SDNFP and DDNFP). Evaluation by immunoblot and dot blot demonstrated the antigenicity and surface presentation of the fused domains (Supplementary Fig. 1). Thermostability evaluation showed that SDNFP and DDNFP could withstand a temperature of ≥60°C without forming aggregates or precipitates (Supplementary Fig. 2).
Immunogenicity and protection of Dps fusion proteins
Various SDNFPs and DDNFPs with antigen domains fused to either or both of N- and C-termini were screened for immunogenicity in mice. Two DDNFPs (M2e-EcDps-FP and M2e-EcDps-CD) were identified by the screening and selected for further evaluation with the SAS adjuvant along with corresponding SDNFPs.
Robust immune responses
DDNFPs induced high levels of antibody responses against both domains (M2e and FP or M2e and CD) with a minimum titer of 5,500 as measured with respective domain peptide as antigens (Fig. 3). Importantly, levels of antibody responses against individual domains by DDNFPs were similar to those by SDNFPs (Fig. 3). These results indicated that the two domains in DDNFPs were independently presented without any apparent interference with each other. They also indicated that Dps was uniquely capable of inducing strong immune response with antigen domains fused separately at N- and C- termini. A strong antibody response to the EcDps carrier was also induced, which was apparently higher than those against individual domains (Fig. 3). This suggests that the EcDps carrier itself is highly immunogenic.
Fig. 3.
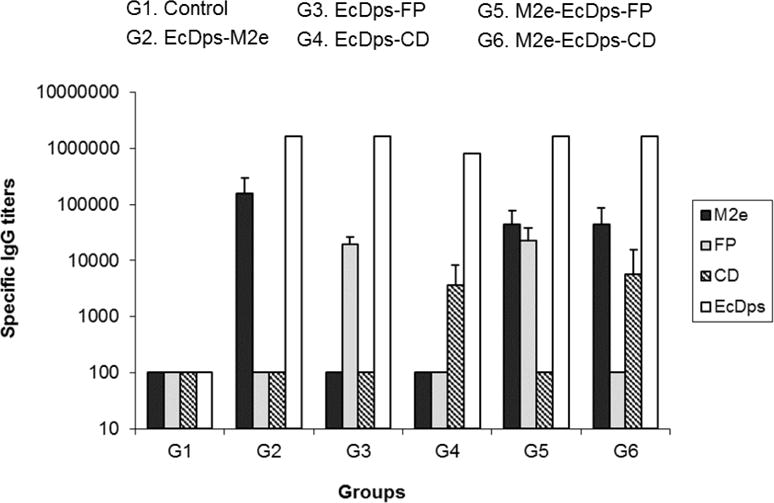
Immunogenicity of SDNFPs and DDNFPs. SDNFPs and DDNSPs were administered to mice (n=5) at 10 μg/dose in combination with SAS twice, 4 weeks apart. Immune sera were tested for specific IgG by ELISA to respective antigen domains at week 2 after the second immunization. The pooled serum sample from each group was used with the carrier protein antigen (EcDps).
Reaction with native antigens
Individual sera generated with SDNFPs and DDNFPs were tested by ELISA using inactivated whole virus antigen of H1N1 NC (A/New Caledonia/20/99). The results showed that each domain induced antibodies reactive with the inactivated whole virus antigen (Fig. 4A). The CD induced a much higher titer than M2e or FP (p<0.05). Correspondingly, the DDNFP with M2e and CD generated a much higher titer than the one with M2e and FP (p<0.05) (Fig. 4A).
Fig. 4.
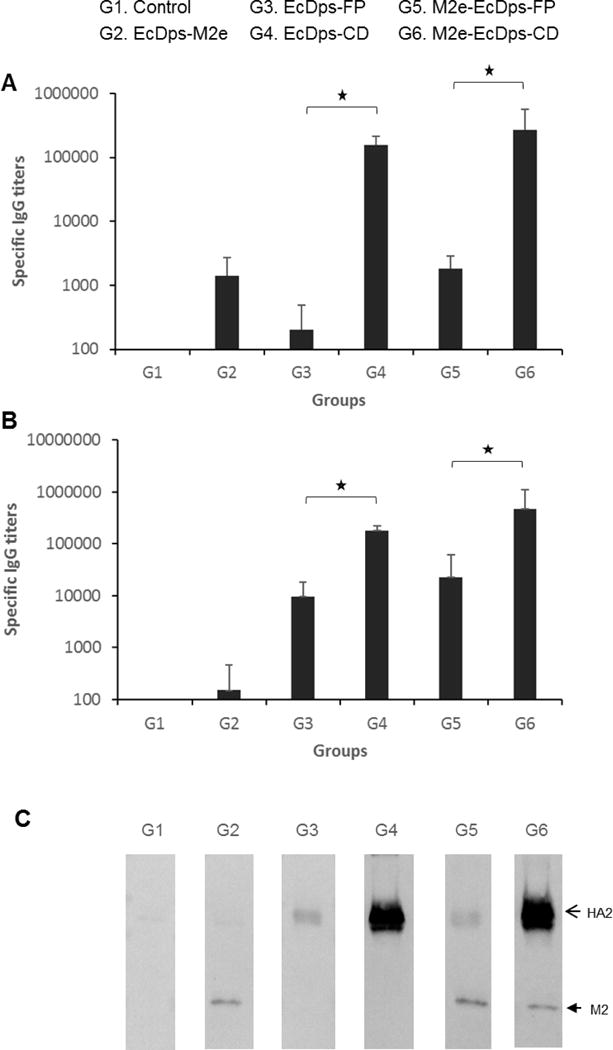
Reaction of specific antibodies induced by the SDNFPs and DDNFPs in mice (n=5) with inactivated whole virus antigen (H1N1 NC) (A) and recombinant HA (H1N1 NC) (B) by ELISA or with inactivated whole virus antigen (H1N1 NC) by immunoblot (C). The open and closed arrows indicate HA2 and M2 proteins, respectively.
The sera were also evaluated by ELISA with recombinant HA (H1N1 NC). The results showed that SDNFPs and DDNFPs containing the FP or CD domain generated high titers of antibodies reactive with recombinant HA (Fig. 4B). As with the inactivated whole virus antigen, the CD domain induced a much higher antibody titer than FP (p<0.05) (Fig. 4B).
Immunoblot was then performed to further confirm the reaction with specific proteins in inactivated whole virus antigen (H1N1 NC). The results showed that antibodies induced by SDNFPs and DDNFPs with M2e, FP and/or CD reacted specifically with the corresponding viral proteins (M2 or HA2) (Fig. 4C). The CD induced a much stronger reaction with HA2 than FP, being consistent with the results by ELISA with inactivated virus or recombinant HA described above (Fig. 4A and 4B). This suggests that the CD domain is more immunogenic than FP. The M2e induced a much weaker reaction (Fig. 4C) as with ELISA against the same antigen (Fig. 4A). This however is likely due in part to the low copy number of M2 per virion (~40) which is about 10 fold lower than that of HA (~ 500) [9,34].
NT and IgG subclasses
Immune sera against SDNFPs and DDNFPs were tested by NT with PR8 (A/Puerto Rico/8/34, H1N1) virus. They did not exhibit a detectable NT titer (<1:20), suggesting that antibodies generated against individual domains or their combination (M2e/FP or M2e/CD) may not be strongly neutralizing and may rely more on other effector mechanisms like ADCC for protection. Analysis of IgG subclasses showed that IgG2a was the dominant subclass with SAS adjuvant as shown with both DDNFPs (Supplementary Fig. 4).
Enhanced protection by domain combination
The protective effect of DDNFPs was evaluated in comparison with SDNFPs. Mice immunized with DDNFPs and SDNFPs as in Fig. 3 were challenged with a highly lethal dose of PR8 virus (5 LD50). All control animals died by days 8–10. Individual domains or SDNFPs (M2e, FP or CD) were independently protective (Fig. 5). The CD domain appeared to be most protective, conferring a complete protection (100% survival), whereas FP was least protective with only a 20% survival (Fig. 5). The M2e provided an 80% survival. This is consistent with the fact that the CD induced much higher antibody titers against native antigens (p<0.05; Fig. 4A).
Fig. 5.
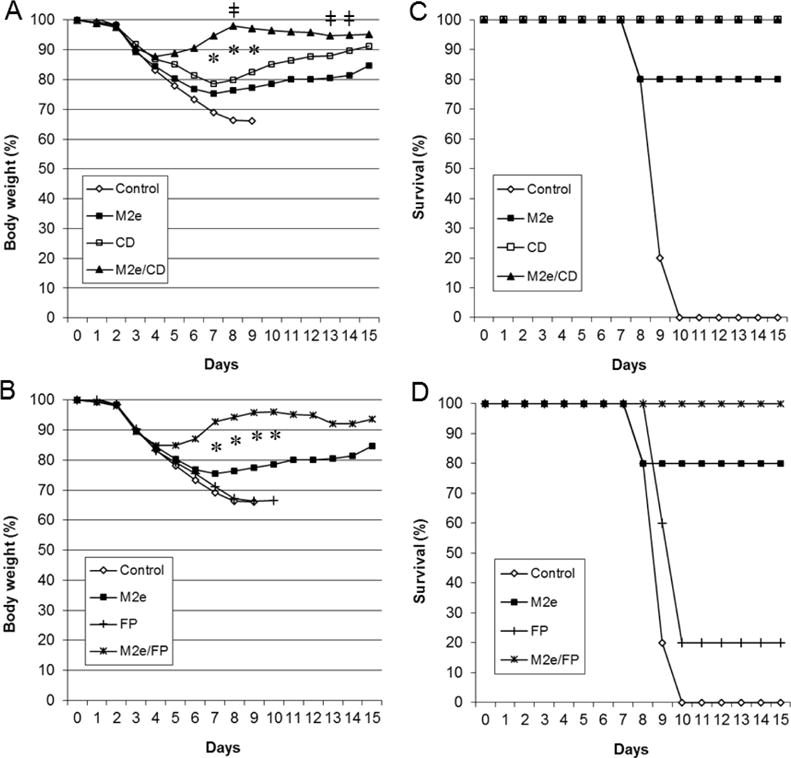
Protective effect of SDNFPs and DDNFPs. Mice (n=5) were challenged with 5 LD50 of H1N1 PR8 at week 3 after the 2nd immunization and monitored for body weight (A and B) and survival (C and D). The data are presented as two separate groups (A and C; B and D) for easy comparison of each DDNFP with its respective SDNFPs. Symbols * indicate significant differences (p<0.05) between DDNFP and either SDNFPs in body weight. Symbols ǂ indicate significant differences (p<0.05) between M2e/CD and M2e/FP DDNFPs in body weight.
The two DDNFPs (M2e-EcDps-FP and M2e-EcDps-CD) provided a greatly enhanced protection as compared to SDNFPs (Fig. 5). They both conferred a complete protection, but with a much smaller body weight loss of 12–15% over only a brief period (days 1–4) and faster body weight recovery (p<0.05) (Fig. 5). In contrast, SDNFPs either failed to provide a complete protection or only generated body weight gain after day 8. Thus, the protection appeared to be synergistically enhanced by combination of two domains (M2e/FP and M2e/CD). The M2e-EcDps-CD exhibited an even stronger protection than M2e-EcDps-FP by reaching significantly higher body weight at some time points during recovery (p<0.05; Fig. 4).
These results were further confirmed in another challenge experiment under the same conditions, but with a higher challenge dose (10 LD50). Only DDNFPs provided a complete protection for survival (p<0.05 by the log-rank test) although with a larger loss of body weight as compared to challenge at 5 LD50 (Fig. 6). In contrast, the survival by SDNFPs was ≤20% (Fig. 6). The M2e-EcDps-CD again provided a better protection than M2e-EcDps-FP based on body weight although the differences were not statistically significant under the conditions used (p>0.05; Fig. 6). These results therefore further demonstrated enhancement of protection by domain combination and the superior protection with the M2e/CD combination.
Fig. 6.
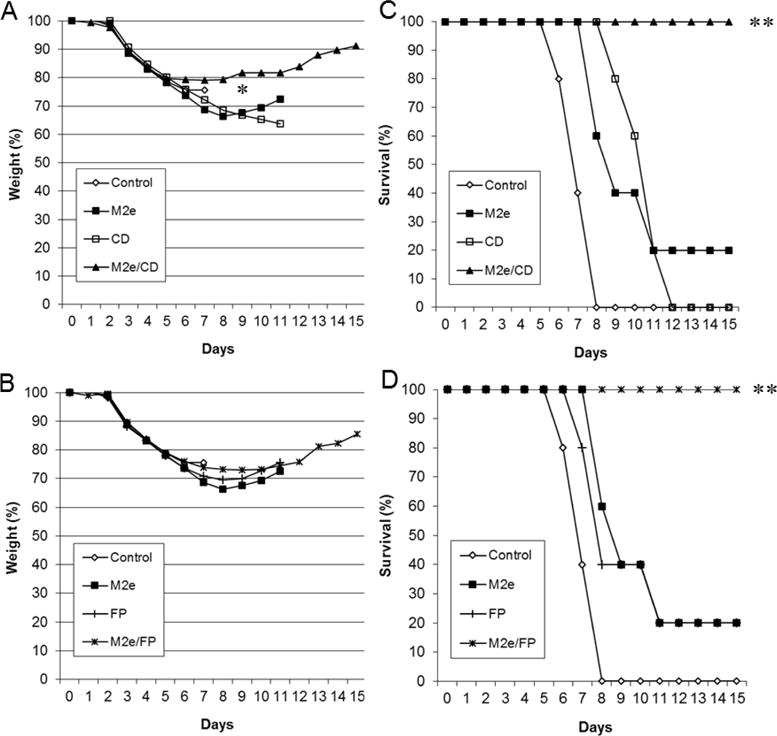
Protective effect of SDNFPs and DDNFPs. Mice (n=5) were challenged with 10 LD50 of H1N1 PR8 at week 3 after the 2nd immunization and monitored for body weight (A and B) and survival (C and D). The data are presented as two separate groups (A and C; B and D) for easy comparison of each DDNFP with its respective SDNFPs. Symbols * indicate significant differences (p<0.05) between DDNFP and either SDNFPs in body weight. Symbols ** indicate significant differences (p<0.05) between DDNFP and either SDNFPs in survival.
Discussion
A potentially highly effective antigen construct (DDNFP) for universal influenza vaccine was generated in the present studies. It is characterized by independent presentation of two conserved domains, synergistically enhanced protection, high-level expression as self-assembled nanoparticles in E. coli, and being thermostable. The DDNFP with M2e and CD domains appeared to be the optimal candidate among different fusion proteins and domains evaluated based on immunogenicity of individual domains as well as protection. To our knowledge, such a DDNFP construct capable of presenting two domains independently and simultaneously has not been previously reported.
The protection by DDNFPs was demonstrated at two different challenge doses (5 and 10 LD50) after a simple two-dose immunization schedule at a moderate antigen dose (two IM injections at 10 μg/mouse with SAS adjuvant). There was only a minimal body weight loss along with full survival when mice were challenged with PR8 at 5 LD50, a challenge dose commonly used with this well-established challenge virus in studies on various universal influenza vaccine candidates. The protection by DDNFPs was clearly synergistically enhanced as compared to the SDNFPs. This is likely due in part to the more efficient killing of viruses by simultaneous binding of antibodies to different viral surface proteins. The DDNFPs did not induce detectable neutralizing antibodies (<1:20) as measured by the standard neutralization assay. This suggests that antibodies induced by DDNFPs against the specific domains (M2e, CD, or FP) may not be strongly neutralizing. These observations are consistent with previous studies with M2e [8,35], while neutralization activity with FP or CD domain did not appear to have been evaluated previously [14–17]. The more sensitive pseudotype neutralization test could be used in future studies to examine the extent of neutralizing activity [5]. On the other hand, strong protection can be mediated by ADCC as shown with M2e [8,35]. Recent studies further showed that ADCC is essential to protection by non-neutralizing as well as neutralizing antibodies [36–37]. Certain IgG subclasses are more effective in mediating ADCC, such as IgG2a in mice [37] which was the dominant subclass induced by DDNFP in combination with SAS adjuvant (Supplementary Fig. 4). Furthermore, a highly protective monoclonal antibody recognizing the stem region of HA was found to be non-neutralizing, but capable of inhibiting virus release [38]. Thus, the ADCC and inhibition of virus release could play a key role in the protection mediated by DDNFPs with these domains.
The conserved domains are usually cryptic or not highly immunogenic as a part of whole protein or antigen [39–40]. The goal of the universal influenza vaccine is therefore to induce high-level immune responses against such domains so that they can be cross-reactive and -protective. The M2e and CD appear to be optimal domains for the DDNFP. Each is from a separate viral surface protein and independently protective and their combination leads to synergistically enhanced protection. It might be even possible to engineer a trimer structure with the CD domain as it is the central part of trimeric stem of HA and the Dps carrier has a 3-fold axis [27–28]. However, even with these conserved domains there could still be variations between groups, clades, and/or species of origin. Thus, M2e is known to vary among different animal species [8]. HA2, while highly conserved within a subtype or closely related subtypes, can vary between clades or groups based on published studies [11,41] as well as those by us (data not shown). Thus, antigen constructs covering different clades or groups may need to be incorporated into the final vaccine composition to achieve a broad-spectrum or universal vaccine. The M2e/CD DDNFP is highly suited for such a multi-component universal influenza vaccine as it is not only highly protective, but can also be produced readily with the bacterial expression system. It thus is suitable for the plug-and-play strategy in developing broadly protective influenza vaccines. We have generated M2e/CD DDNFPs representing other clades or groups, including H3 (group 2), with the goal of establishing the core vaccine composition as the final vaccine product candidate.
Supplementary Material
Acknowledgments
This work was supported in part by a SBIR grant (R43AI092923) from National Institute of Allergy and Infectious Diseases (NIAID), National Institute of Health (NIH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Potential conflicts of interest
The authors declare no conflicts of interest.
References
- 1.Zhang H, Wang L, Compans RW, Wang BZ. Universal influenza vaccines, a dream to be realized soon. Viruses. 2014;6:1974–1991. doi: 10.3390/v6051974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nabel GJ, Fauci AS. Induction of unnatural immunity: prospects for a broadly protective universal influenza vaccine. Nat Med. 2010;16:1389–91. doi: 10.1038/nm1210-1389. [DOI] [PubMed] [Google Scholar]
- 3.Pica N, Palese P. Toward a universal influenza virus vaccine: prospects and challenges. Annu Rev Med. 2013;64:189–202. doi: 10.1146/annurev-med-120611-145115. [DOI] [PubMed] [Google Scholar]
- 4.Impagliazzo A, Milder F, Kuipers H, Wagner MV, Zhu X, Hoffman RM, van Meersbergen R, Huizingh J, Wanningen P, Verspuij J, de Man M, Ding Z, Apetri A, Kükrer B, Sneekes-Vriese E, Tomkiewicz D, Laursen NS, Lee PS, Zakrzewska A, Dekking L, Tolboom J, Tettero L, van Meerten S, Yu W, Koudstaal W, Goudsmit J, Ward AB, Meijberg W, Wilson IA, Radošević K. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science. 2015;349:1301–6. doi: 10.1126/science.aac7263. [DOI] [PubMed] [Google Scholar]
- 5.Yassine HM, Boyington JC, McTamney PM, Wei CJ, Kanekiyo M, Kong WP, Gallagher JR, Wang L, Zhang Y, Joyce MG, Lingwood D, Moin SM, Andersen H, Okuno Y, Rao SS, Harris AK, Kwong PD, Mascola JR, Nabel GJ, Graham BS. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat Med. 2015;21:1065–1070. doi: 10.1038/nm.3927. [DOI] [PubMed] [Google Scholar]
- 6.Wohlbold TJ, Nachbagauer R, Margine I, Tan GS, Hirsh A, Krammer F. Vaccination with soluble headless hemagglutinin protects mice from challenge with divergent influenza viruses. Vaccine. 2015;33:3314–21. doi: 10.1016/j.vaccine.2015.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du L, Zhou Y, Jiang S. Research and development of universal influenza vaccines. Microbes Infect. 2010;12:280–6. doi: 10.1016/j.micinf.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 8.Deng L, Cho KJ, Fiers W, Saelens X. M2e-Based Universal Influenza A Vaccines. Vaccines (Basel) 2015;3:105–36. doi: 10.3390/vaccines3010105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zebedee SL, Lamb RA. Influenza virus M2 protein: monoclonal antibody restriction of virus growth and detection of M2 in virions. J Viral. 1988;62:2762–2772. doi: 10.1128/jvi.62.8.2762-2772.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 11.Fouchier RA, Munster V, Wallensten A, Butterbeer TM, Herfst S, Smith D, Rimmelzwaan GF, Olsen B, Osterhaus AD. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J Virol. 2005;79:2814–2822. doi: 10.1128/JVI.79.5.2814-2822.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steel J, Lowen AC, Wang TT, Yondola M, Gao Q, Haye K, García-Sastre A, Palese P. Influenza virus vaccine based on the conserved hemagglutinin stalk domain. MBio. 2010;18:1. doi: 10.1128/mBio.00018-10. 2010. pii: e00018-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chun S, Li C, Van Domselaar G, Wang J, Farnsworth A, Cui X, Rode H, Cyr TD, He R, Li X. Universal antibodies and their applications to the quantitative determination of virtually all subtypes of the influenza A viral hemagglutinins. Vaccine. 2008;26:6068–76. doi: 10.1016/j.vaccine.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 14.Staneková Z, Király J, Stropkovská A, Moskva T, Micha V, Kostolanský F, Barkov E. Heterosubtypic protective immunity against influenza A virus induced by fusion peptide of the hemagglutinin in comparison to ectodomain of M2 protein. Acta Virol. 2011;55:61–67. doi: 10.4149/av_2011_01_61. [DOI] [PubMed] [Google Scholar]
- 15.Wang TT, Tan GS, Hai R, Pica N, Ngai L, Ekiert DC, Wilson IA, García-Sastre A, Moran TM, Palese P. Vaccination with a synthetic peptide from the influenza virus hemagglutinin provides protection against distinct viral subtypes. Proc Natl Acad Sci U S A. 2010;107:18979–84. doi: 10.1073/pnas.1013387107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xin L, Yang XY, Yu ZJ, Bo H, Zhou JF, Qin K, Shu YL. Preliminary study of a universal vaccine based on the HA2 protein of the H5N1 influenza virus. Bing Du Xue Bao. 2014;30:521–528. [PubMed] [Google Scholar]
- 17.Chen S, Zheng D, Li C, Zhang W, Xu W, Liu X, Fang F, Chen Z. Protection against multiple subtypes of influenza viruses by virus-like particle vaccines based on a hemagglutinin conserved epitope. Biomed Res Int. 2015;90:1817. doi: 10.1155/2015/901817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee YT, Kim KH, Ko EJ, Lee YN, Kim MC, Kwon YM, Tang Y, Cho MK, Lee YJ, Kang SM. New vaccines against influenza virus. Clin Exp Vaccine Res. 2014;3:12–28. doi: 10.7774/cevr.2014.3.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neirynck S, Deroo T, Saelens X, Vanlandschoot P, Joy WM, Fiers W. A universal influenza A vaccine based on the extracellular domain of the M2 protein. Nat Med. 1999;5:1157–63. doi: 10.1038/13484. [DOI] [PubMed] [Google Scholar]
- 20.Denis J, Acosta-Ramirez E, Zhao Y, Hamelin ME, Koukavica I, Baz M, Abed Y, Savard C, Pare C, Lopez Macias C, Boivin G, Leclerc D. Development of a universal influenza A vaccine based on the M2e peptide fused to the papaya mosaic virus (Pump) vaccine platform. Vaccine. 2008;26:3395–403. doi: 10.1016/j.vaccine.2008.04.052. [DOI] [PubMed] [Google Scholar]
- 21.Wang W, Huang B, Jiang T, Wang X, Qi X, Gao Y, Tan W, Ruan L. Robust immunity and heterologous protection against influenza in mice elicited by a novel recombinant NP-M2e fusion protein expressed in E. coli. PLoS One. 2012;7:e52488. doi: 10.1371/journal.pone.0052488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X, Guo J, Han S, Yao L, Chen A, Yang Q, Bo H, Xu P, Yin J, Zhang Z. Enhanced immune response induced by a potential influenza A vaccine based on branched M2e polypeptides linked to tuftsin. Vaccine. 2012;30:6527–33. doi: 10.1016/j.vaccine.2012.08.054. [DOI] [PubMed] [Google Scholar]
- 23.Huleatt JW, Nakaar V, Desai P, Huang Y, Hewitt D, Jacobs A, Tang J, McDonald W, Song L, Evans RK, Umlauf S, Tussle L, Powell TJ. Potent immunogenicity and efficacy of a universal influenza vaccine candidate comprising a recombinant fusion protein linking influenza M2e to the TLR5 ligand flagellin. Vaccine. 2008;26:201–14. doi: 10.1016/j.vaccine.2007.10.062. [DOI] [PubMed] [Google Scholar]
- 24.Hashemi H, Pouyanfard S, Bandehpour M, Noroozbabaei Z, Kizami B, Saelens X, Mokhtari-Azad T. Immunization with M2e-displaying T7 bacteriophage nanoparticles protects against influenza A virus challenge. PLoS One. 2012;7:e45765. doi: 10.1371/journal.pone.0045765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou D, Wu TL, Lasaro MO, Latimer BP, Parzych EM, Bian A, Li Y, Li H, Erikson J, Xiang Z, Ertl HC. A universal influenza A vaccine based on adenovirus expressing matrix-2 ectodomain and nucleoprotein protects mice from lethal challenge. Mol Ther. 2010;18:2182–9. doi: 10.1038/mt.2010.202. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L, Wang YC, Feng H, Ahmed T, Compans RW, Wang BZ. Virus-like particles containing the tetrameric ectodomain of influenza matrix protein 2 and flagellin induce heterosubtypic protection in mice. Biomed Res Int. 2013;2013:686549. doi: 10.1155/2013/686549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grant RA, Filman DJ, Finkel SE, Kolter R, Hogle JM. The crystal structure of Dps, a ferritin homolog that binds and protects DNA. Nat Struct Biol. 1998;5:294–303. doi: 10.1038/nsb0498-294. [DOI] [PubMed] [Google Scholar]
- 28.Haikarainen T, Papageorgiou AC. Dps-like proteins: structural and functional insights into a versatile protein family. Cell Mol Life Sci. 2010;67:341–51. doi: 10.1007/s00018-009-0168-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barrett T, Angliss SC. Growth, purification, and titration of influenza viruses. In: Many WJ, editor. Virology-A Practical Approach. IRL press; Oxford: 1985. pp. 119–150. [Google Scholar]
- 30.WHO. (Technical Report Series, No. 927).Recommendations for the production and control of influenza vaccine (inactivated) 2005 Available at http://www.wpro.who.int.
- 31.WHO/CDS/CSR/NCS/2002.5. Manual on animal influenza diagnosis and surveillance Department of communicable disease surveillance and response. World Health Organization Global Influenza Program; Available at http://www.wpro.who.int. [Google Scholar]
- 32.Bewick V, Cheek L, Ball J. Statistics review 12: survival analysis. Crit Care. 2004;8:389–94. doi: 10.1186/cc2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruth A, Gliotic M, Nadeau VG, Chen G, Deber CM. Detergent binding explains anomalous SDS-PAGE migration of membrane proteins. Proc Natl Acad Sci U S A. 2009;106:1760–5. doi: 10.1073/pnas.0813167106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lamb RA, Krug RM. Orthomyxoviridae: the viruses and their replication. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. Fields virology. 4th. Lippincott Williams & Wilkins; Philadelphia, Pa: 2001. pp. 1487–1531. [Google Scholar]
- 35.Jegerlehner A, Schmitz N, Stormy T, Bachmann MF. Influenza A vaccine based on the extracellular domain of M2: weak protection mediated via antibody-dependent NK cell activity. J Immunol. 2004;172:5598–605. doi: 10.4049/jimmunol.172.9.5598. [DOI] [PubMed] [Google Scholar]
- 36.Jegaskanda S, Job ER, Kramski M, Laurie K, Isitman G, de Rose R, Winnall WR, Stratov I, Brooks AG, Reading PC, Kent SJ. Cross-reactive influenza-specific antibody-dependent cellular cytotoxicity antibodies in the absence of neutralizing antibodies. J Immunol. 2013;190:1837–48. doi: 10.4049/jimmunol.1201574. [DOI] [PubMed] [Google Scholar]
- 37.Vanderven HA, Jegaskanda S, Wheatley AK, Kent SJ. Antibody-dependent cellular cytotoxicity and influenza virus. Curd Opsin Virol. 2017;22:89–96. doi: 10.1016/j.coviro.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 38.Yamayoshi S, Uralic R, Ito M, Kiso M, Makutus S, Yasuhara A, Oishi K, Sasaki T, Ikuta K, Kawaoka Y. A broadly reactive human anti-hemagglutinin stem monoclonal antibody that inhibits influenza A virus particle release. EBioMedicine. 2017 doi: 10.1016/j.ebiom.2017.03.007. Pio. S2352-3964(17)30094-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gerhard W, Mozdzanowska K, Zharikova D. Prospects for universal influenza virus vaccine. EMerge Infect Dis. 2006;12:569–74. doi: 10.3201/eid1204.051020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sui J, Sheehan J, Hwang WC, Bankston LA, Burchett SK, Huang CY, Liddington RC, Beiges JH, Marasco WA. Wide prevalence of heterosubtypic broadly neutralizing human anti-influenza A antibodies. Clin Infect Dis. 2011;52:1003–09. doi: 10.1093/cid/cir121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fan X, Hashem AM, Chen Z, Li C, Doyle T, Zhang Y, Yi Y, Farnsworth A, Xu K, Li Z, He R, Li X, Wang J. Targeting the HA2 subunit of influenza A virus hemagglutinin via CD40L provides universal protection against diverse subtypes. Mucosal Immunology. 2015;8:211–20. doi: 10.1038/mi.2014.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


