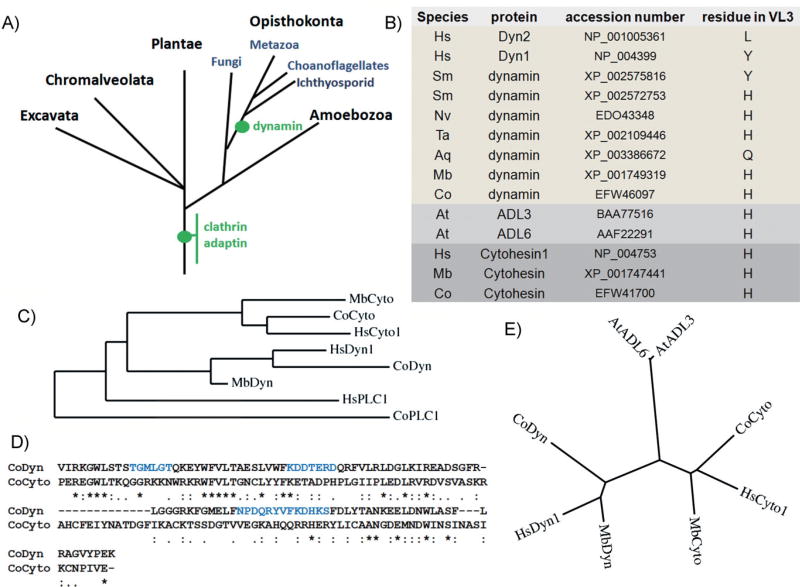
Figure 2. Evolution of the dynamin PHD.
A: Model for major acquisition of selected proteins in eukaryotes. The evolutionary relationships of various eukaryotic taxa are shown [8], and the positions of acquisitions of gene families are indicated (green dots). B: Table showing the identity of the diagnostic residue in VL3 in dynamins from various organisms. In lower organisms the residue is a histidine, which is conserved in the VL3 of the related PHD from cytohesin (ArfGEF). C: A common ancestor for PHDs of cytohesins and dynamins. PHDs from dynamins, cytohesins, and phospholipase Cγ 1 (PLC1) are aligned and plotted as the rooted phylogenetic tree. D: Alignment of PHDs from an early premetazoan organism C. owczarzaki (Co). Shown is the PHD from dynamin and cytohesin. PHDs are aligned with CLUSTAL 2.0.1, and the VLs are colored in blue. Asterisk indicates identical amino acid, and period or colon represent similar amino acids. E: Unrooted phylogenetic tree of PHDs from dynamins, cytohesins, and Arabidopsis dynamin-like proteins, ADL3 and ADL6, is plotted with Phylogeny.fr. The abbreviation of organisms include Homo sapiens (Hs), S. mansoni (Sm), Nematostella vectensis (Nv), Trichoplax adhaerens (Ta), Amphimedon queenslandica (Aq), M. brevicollis (Mb), C. owczarzaki (Co), and A. thaliana (At).