Abstract
Twenty years ago, chromosomal abnormalities were the only identifiable genetic causes of a small fraction of congenital heart defects (CHD). Today, a de novo or inherited genetic abnormality can be identified as pathogenic in one-third of cases. We refer to them here as monogenic causes, insofar as the genetic abnormality has a readily detectable, large effect. What explains the other two-thirds? This review considers a complex genetic basis. That is, a combination of genetic mutations or variants that individually may have little or no detectable effect contribute to the pathogenesis of a heart defect. Genes in the embryo that act directly in cardiac developmental pathways have received the most attention, but genes in the mother that establish the gestational milieu via pathways related to metabolism and aging also have an effect. A growing body of evidence highlights the pathogenic significance of genetic interactions in the embryo and maternal effects that have a genetic basis. The investigation of CHD as guided by a complex genetic model could help estimate risk more precisely and logically lead to a means of prevention.
Keywords: Congenital heart defects, Genetics, Maternal age, Maternal effects, Modifier gene
Gregor Mendel presented the laws of inheritance at a meeting of the Natural History Society of Brünn in 1865. The mathematics in his talk lost the audience, and the accompanying paper was soon forgotten.1 In 1866, Thomas Peacock described a child with tetralogy of Fallot whose sibling had “something the matter with its heart.” He speculated that “strong mental impressions or shocks which were sustained during pregnancy” caused cardiac malformation in some cases and a “hereditary predisposition” in others.2 Albeit vague, the notion that congenital heart defects (CHD) have a genetic basis predated the rediscovery of Mendel’s laws in the 1900 s.
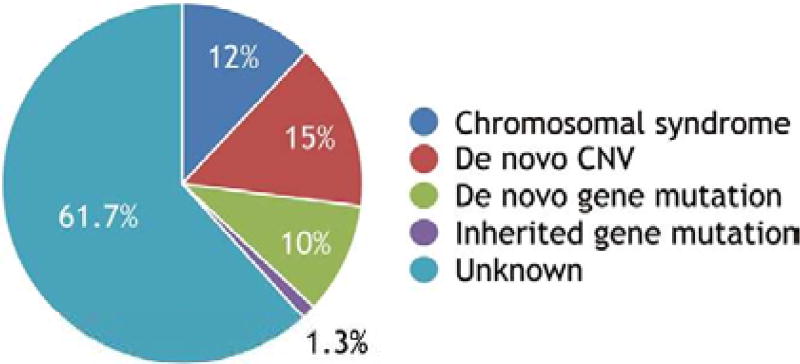
Of course, most CHD occur sporadically and do not fit simple Mendelian patterns of inheritance, which led James Nora to propose in 1968 that a combination of genetic and environmental factors contribute to the development of a heart defect in an individual. The model appealed to him because it suggested that multiple factors could be targeted to prevent CHD.3 Nora’s reasoning was elegant, but the multifactorial hypothesis was difficult to validate when no etiology was known in the vast majority of cases. Investigators accordingly designed studies to discover causes that have readily detectable, large effects. Genectic abnormalities in the embryo comprise the vast majority of the known causes. For the sake of discussion, we refer to a genetic abnormality as monogenic if it can be considered the cause of a heart defect. The abnormality may involve a single gene, chromosomal interval or entire chromosome. As discussed later, other factors can modify the risk of a heart defect in the presence of the monogenic cause. The modifiers do not necessarily cause disease on their own. Currrently, a monogenic basis can be identified in one-third of all cases of CHD (figure).4–9
Figure.
The vast majority of the known causes of congenital heart defects (CHD) are de novo or inherited genetic abnormalities. A monogenic basis can be identified in one-third of all cases. The Baltimore-Washington Infant Study and Metropolitan Atlanta Congenital Defects Program yielded similar estimates for the fraction attributable to chromosomal syndromes.4,5 De novo copy number variants (CNVs) include both the well-known, such as the 22q11.2 deletion, and more recently discovered ones.6 The fraction attributable to de novo mutations that affect protein-coding sequence were recently reported in 3 large, whole-exome sequencing studies of trios.7–9 The burden of inherited loss-of-function mutations of known CHD genes was estimated in one of these studies.9 Inherited CNVs and other forms of genetic mutation cause monogenic CHD, but their attributable fractions have not been quantified in large studies similar to the ones cited above. An oligogenic basis may explain a large fraction of the currently unknown causes.
De novo genetic abnormalities can explain a significant fraction of CHD, but are not the entire explanation. Epidemiologic studies have consistently supported a significant role for inheritance.10,11 The largest study, encompassing >1.7 million persons born in Denmark between 1977 and 2005, showed that a family history of any CHD is a strong risk factor.10 Given an affected first-degree relative, the relative risk (RR) is more than 3-fold higher after excluding cases of chromosomal anomalies or extracardiac defects. The risk falls with the degree of distance from the proband but remains significant to third-degree relations. Few risk factors are as strong or consistent. A couple of notable exceptions include maternal phenylketonuria, which increases the risk 6-fold, and pregestational diabetes, which has comparable risk.12–15
The fraction of cases that can be attributed to an inherited mutation is more difficult to estimate because of incomplete penetrance, unrecognized pathogenic mutations and undiscovered CHD genes. A recent whole-exome sequencing study of trios (i.e., the affected child and parents) offers one estimate.9 Non-syndromic patients with an isolated CHD are more likely to have inherited mutations of a known CHD gene or other genes. An inherited protein-truncating variant of a CHD gene was identified in a significant, but small fraction of non-syndromic patients (1.3%: 17/1,281; figure). This fraction is a conservative estimate because it is limited to loss-of-function mutations of known CHD genes. The same study also found a significant excess (3,318) of protein-truncating variants of other genes that are likely to have undiscovered functions in cardiac development. Finally, there was a trend towards an excess of missense mutations of CHD genes (163 variants, P=0.0863). The number of patients who had these other mutations was not given, but tallying the results suggests that a patient with isolated CHD has an average of approximately 2.73 excess, inherited mutations. A pathogenic combination of inherited mutations could explain a large fraction of cases that currently have an unknown cause.
Monogenic CHD: One End of the Complex Genetic Continuum
Viewing CHD as either monogenic or complex is as much a consequence of experimental design as biology. In a typical human study, cases are selected for the presence of a CHD, and the goal is to identify 1 highly penetrant genetic abnormality in each case. For instance, multiplex families are studied, or de novo mutations are sought in trios of unaffected parents and affected child. The high penetrance of a monogenic cause is demonstrated by its segregation with affected relatives or its enrichment in cases over controls. The identification of 1 genetic cause, however, does not exclude the contribution of other genes to the pathogenesis of each case.
When studies ascertain cases by genetic diagnosis rather than CHD, well-established monogenic causes consistently show high, but incomplete penetrance. The pattern is the same whether the cause is a chromosomal anomaly that affects more than 1 gene or the mutation of a single gene (Table l).16–25 Additional factors modify the risk of CHD caused by a single genetic abnormality. Hearkening to Nora’s multifactorial hypothesis, investigators have sought to identify the other contributing environmental or genetic factors. Not for lack of effort, only a few environmental factors have been clearly and reproducibly demonstrated to cause disease or to affect the risk of CHD in general.15 Similarly, it has been difficult to establish the role of environmental factors in monogenic CHD. For example, maternal folate supplementation, which has received much attention in Down syndrome, has no or little detectable effect.26,27 In contrast, the evidence for modifier genes is growing in humans and well established in mouse models.
Table 1.
Genetic Causes of CHD
| Species / Genetic abnormality | CHD incidence, % |
Ref. |
|---|---|---|
| Human | ||
|
| ||
| 1q21.1 deletion | 23 | 16 |
|
| ||
| Turner syndrome (45, XO karyotype) | 30 | 17 |
|
| ||
| Cornelia de Lange (NIPBL, SMC1A, SMC3) | 31 | 18 |
|
| ||
| Down syndrome (trisomy 21) | 44 | 19 |
|
| ||
| Holt-Oram syndrome (TBX5) | 62 | 20 |
|
| ||
| DiGeorge syndrome (22q11.2 deletion) | 75 | 21 |
|
| ||
| NKX2–5 mutation | 88 | 22 |
|
| ||
| Mouse | ||
|
| ||
| Gata4+/− | 12–76 | 23 |
|
| ||
| Nkx2–5+/− | 5–50 | 24 |
|
| ||
| Tbx5+/− | 40–80 | 25 |
Genetic causes of CHD show high, but never complete penetrance. Examples of human copy number variants, chromosomal, and genetic abnormalities are shown in which cases were ascertained by the presence of the genetic abnormality. CHD incidence in mouse mutant models depends on the genetic background, which can be systematically varied via inbred strain crosses. The variability is a sign of interactions between the mutated CHD gene and polymorphic modifier genes.
CHD, congenital heart defects.
Consider the familial recurrence risk of CHD. The risk is commonly presumed to be from monogenic causes that are private to families, but it could also be caused by transmission of genetic modifiers that affect risk independently of the cause in a particular relative. Two groups have considered the latter hypothesis in the setting of CHD caused by a 22q11.2 deletion in the proband.28,29 Their studies yielded consistent results with a combined total of 212 probands and almost 2,000 first- and second-degree relatives who did not have the deletion. In the larger of the 2 studies, the incidence of CHD in the relatives was 4-fold greater if the proband had CHD than if not. Moreover, the incidence of severe CHD in the relatives of probands who had CHD was 6-fold greater than in the general population.29 More studies are necessary to determine whether the familial recurrence risk is independent of a monogenic cause in the proband.
Common genetic variation, either common copy number variants (CNVs) or single nucleotide polymorphisms, has not been found to have a detectable risk on monogenic CHD, with one telling exception.30–32 A 12p13.31 CNV duplication is associated with the risk of conotruncal heart defects and left-sided lesions in DiGeorge and Turner syndromes, respectively.30,33 The interval includes 3 genes, SLC2A3, SLC2A14, and NANOGP1. The functions of SLC2A14 and NANOGP1 are unknown. SLC2A14 encodes GLUT14, one of a large class of proteins with homology to facilitative glucose transporters. NANOGP1 is a pseudogene of NANOG, the transcription factor that maintains pluripotency. SLC2A3 encodes GLUT3, a facilitative glucose transporter that is expressed by trophoblasts in the placenta. GLUTs facilitate the transport of glucose across cell membranes and down a concentration gradient. Haploinsufficiency of SLC2A3 causes a quantitative reduction in placental glucose transport and embryonic glucose concentration.34 One may speculate that maternal diabetes and a SLC2A3 duplication similarly increase the risk of CHD by causing embryonic hyperglycemia. Maternal diabetes would do so by increasing the glucose concentration gradient between the mother and embryo, while the SLC2A3 duplication would increase glucose transport into the embryo. The RR associated with maternal diabetes in a recent Danish epidemiologic study (RR 4.00, 95% confidence interval (CI) 3.51–4.53) was modestly higher than the risk associated with the SLC2A3 duplication in DiGeorge (RR 1.4; 95% CI 1.3–1.6) and Turner syndromes (RR 1.8; 95% CI 1.3–2.5. The RRs for each syndrome were recalculated from the published data for comparison.)13,30,33 The risk associated with maternal diabetes may be higher because maternal hyperglycemia can induce greater embryonic hyperglycemia than can a SLC2A3 duplication when the mother is normoglycemic.
Rare genetic variation of cardiac developmental genes has been found to affect the risk of monogenic CHD. For example, mutations of CRELD1 and HEY2 are known to cause CHD in humans and mice, respectively.35,36 Rare variants of these 2 genes have also been associated with atrioventricular septal defects (AVSD) in Down syndrome. Genetic experiments in a mouse model of human trisomy 21 support the findings.37,38 Similarly, mutations of histone-modifying genes are an important class of monogenic causes of CHD.7 In a whole-exome sequencing study of DiGeorge syndrome patients, rare variants of several histone-modifying genes were associated with an increased risk of conotruncal heart defects. Conversely, rare variants of genes that have the opposite biochemical effect on histone modifers fication may be associated with a reduced risk.39
The major role of modifier genes in monogenic CHD has been demonstrated in mouse models via 1 of 2 experimental strategies. The most common is to examine the interaction between a CHD gene and a candidate modifier gene. Typically, 2 mutant lines that each carry a knockout mutation of the CHD or candidate gene are crossed. A difference in the incidences of CHD between the 2 single and the double mutant progeny indicates a genetic interaction, as has been shown for several monogenic CHD models (Table 2).40–50 Many more pathologically significant genetic interactions undoubtedly remain to be discovered.
Table 2.
Genetic Interactions Between a CHD Gene and a Modifier Gene
| CHD gene | Modifier gene | Ref. |
|---|---|---|
| Gata4 | Gata5*, Tbx5* | 40, 41 |
| Jag1 | Notch2* | 42 |
| Nkx2–5 | Smad1, Nipbl*, Whsc1 | 43–45 |
| Tbx1 | Crkl*, Eya1, Fgf8, Prdm1 | 46–50 |
Genetic interactions between a CHD gene and a modifier gene affect the expression of monogenic phenotypes. Examples are shown in which crosses between mouse knockouts of a human CHD gene and a candidate modifier gene produced double mutants that had an exacerbated mutant phenotype with one exception. Smad1 loss-of-function actually rescued the outflow tract defects of Nkx2–5 mutant embryos.
Known human CHD gene.
CHD, congenital heart defects.
The second strategy demonstrates the effects of naturally occurring genetic polymorphisms that interact with the disease-causing mutation. Mouse mutants are usually studied in a homogeneous, inbred strain background so that phenotypes can be clearly attributed to the mutated gene. In contrast, a few groups have characterized monogenic CHD models in heterogeneous genetic backgrounds. Introduction of the mutation into systematic inbred strain crosses permits the analysis of genetic interactions with polymorphic modifier genes. Regardless of the particular mutation, the incidence of CHD varies widely with the genetic background (Table 1). In a study of >3,000 Nkx2–5+/− mice from 5 different crosses, the incidence of specific malformations, such as membranous ventricular septal defect (VSD) or AVSD, also varied with the genetic background.24 Alleles of modifier genes either increase or decrease the susceptibility of a cardiac developmental pathway to the causative mutation. Combinations of low- and high-risk genotypes at quantitative trait loci (QTL) determine the risk of specific cardiac malformations in the Nkx2–5+/− mouse. Several QTLs for VSDs have been described from a cross between 2 inbred strains.51 An ongoing effort in our group has yielded at least a dozen QTLs for simple and complex heart defects.
Evidence for the Oligogenic Basis of Human CHD
Although a monogenic cause of CHD is defined by its high phenotypic penetrance in a population, its interaction with modifier genes can significantly promote or suppress risk in an individual. Genetic interactions can likewise potentiate the deleteriousness of a mutation that has little or no effect on its own. Oligogenic combinations of inherited genetic variants could explain the majority of CHD cases that lack a detectable monogenic basis.
Early studies noted that affected individuals not uncommonly carry rare, heterozygous mutations of 2 different CHD genes that are predicted to be deleterious.52–54 The studies were limited by DNA-sequencing capacity, so only a few genes were examined in a small number of individuals. Two recent studies suggest that oligogenic combinations of mutations are a common pathogenic phenomenon.
Using whole-exome sequencing, the first study examined individuals with AVSDs caused by mutations of known AVSD genes. The cases carried more, presumably pathogenic mutations of AVSD genes per person than the general population.55 On average, each case and control respectively carried 3.62 and 2.40 rare, nonsynonymous mutations and 1.70 and 1.36 rare, nonsynonymous, and predicted damaging mutations of an AVSD gene. The number of rare, synonymous mutations, which should not affect gene function, was the same in cases and controls (1.78 and 1.76, respectively; n=81 cases and 4,300 population controls; averages were recalculated from the published data). Interestingly, rare and novel nonsynonymous variants were significantly enriched in 6 genes: NIPBL, CHD7, CEP152, BMPR1a, ZFPM2, and MDM4. Of 34 probands who had mutations of these genes, 8 had a rare or rare, damaging mutation of >1 of the genes. The quantitative differences in the numbers of mutations per case and control suggest that an oligogenic combination of mutations act additively or synergistically to cause disease.
The second study experimentally validated an oligogenic basis of heterotaxy-associated CHD.56 Monogenic mutations that perturb the structure or function of cilia cause CHD, including cases not associated with heterotaxy.7,57 Sequencing approximately 900 ciliome genes, the investigators discovered 8 heterozygous mutations of DNAH6 among 162 cases; 6 of the cases also carried a rare, heterozygous mutation of at least 1 other ciliome gene: DNAI1 in 1 case and DNAH5 in 4 others. Mutations of DNAI1 are known to cause primary ciliary dyskinesia. In zebrafish and mouse models, a modest reduction in the gene dosage of DNAH6, DNAI1, or DNAH5 individually had little or no effect, whereas the same reduction of the genes in pairs (e.g., DNAH6 and DNAI1 or DNAH6 and DNAH5) synergistically increased the incidence of left-right patterning defects, abnormal heart looping and ciliary dysfunction.
Maternal Genes Affecting the Gestational Milieu Contribute to the Genetic Complexity
Genetic mutations in the embryo are the most well-recognized cause of abnormal cardiac development. Maternal genetic factors that foster an adverse gestational milieu are less well studied, yet they may offer the best opportunities for reducing the risk of CHD.58 After all, adult conditions are more easily treated than mutations in the embryo. For example, maternal mutations of phenylalanine hydroxylase, which cause phenylketonuria, or genetic polymorphisms that contribute to pregestational diabetes can indirectly increase the risk of CHD, but proper medical management can reduce the risk.
Older maternal age is also a risk factor for CHD.59–65 The risk was found as well in the Nkx2–5+/− mouse model, which offered the opportunity to characterize the mechanism.51,66 Conventional wisdom ascribes the basis of the risk to the oocyte. A reciprocal ovarian transplant experiment between young and old mothers revealed, however, that the incidence of CHD in Nkx2–5+/− offspring is related to the age of the mother and not the ovary. Maternal age did not affect risk for the wildtype. Maternal hyperglycemia or obesity, both of which are associated with aging and human CHD, does not explain the mechanism because a high-fat diet did not exacerbate the risk. Most interestingly, voluntary exercise by mothers reduced the risk for the Nkx2–5+/− offspring of old mothers by half. The absolute incidence, approximately 10% for VSDs, was equivalent to that in the offspring of young mothers. Aging is not a disease per se, but there is an associated condition that can potentially be managed in mothers to reduce CHD risk in the offspring.
The maternal age-associated risk varied quantitatively between inbred strain crosses of mice.66 Maternal genes determine the activity of a putative age-related factor that interacts with cardiac development in Nkx2–5+/− embryos. A broad perspective on the complex genetic basis of CHD should consider not only genes expressed in the embryo, but also the genes in the mother that establish the gestational milieu for embryonic cardiac development.
Future Directions
Mutations of approximately 200 genes are associated with human CHD.9 The mutations mainly affect transcription factors, epigenetic regulators, and signaling pathways. Tremendous progress has been made in understanding how the mutations affect cardiac development. The results have found relevance to clinical problems such as arrhythmia and cardiomyopathy,67 but how the knowledge can be applied to prevent CHD is far less clear. Nevertheless, we believe that CHD prevention should remain a driving force for research. With that in mind we outline 3 research areas that could yield novel methods to quantify and reduce the risk of CHD.
Contributions to the Incomplete Penetrance of Monogenic CHD Mutations
Mutations that cause severe CHD should be subject to negative selection. Indeed, our analysis of data from the Exome Aggregation Consortium (ExAC)68 indicates that humans are more intolerant of loss-of-function mutations of CHD genes than all other genes (pLI scores 0.59±0.03 vs. 0.30±0.003, mean ± SEM; P=3×10−22. The supplementary table 20 in Sifrim et al9 lists the CHD genes.) On the other hand, 1.9% of adults in the ExAC study, who did not have any severe pediatric disease, carry a loss-of-function mutation of a CHD gene. Given that the incidence of moderate-to-severe CHD at birth is 0.6%,69 there may be genetic mechanisms that suppress the deleteriousness of mutations, as has been observed in mouse models.24,25 Delineation of the genetic modifiers could help to provide more personalized estimates of familial recurrence risks that are currently based on epidemiologic data.
Genetic Interactions that Contribute to the Oligogenic Basis of CHD
The interactions between genetic variants may be just as important as individual genes in the pathogenesis of a large fraction of cases. Guided by the monogenic model, large whole-exome sequencing studies of trios have focused on high-heart expressed or known CHD genes and loss-of-function mutations.7–9 In contrast, statistical and bioinformatic analyses of human genomic studies are much more difficult when there are exponentially more interactions than genes to consider and the interacting genetic variants cannot be as simply or narrowly defined as a loss-of-function mutation. Systems genetic analysis in animal models could help to narrow the search space for human studies. If synergistic interactions contribute significantly to the pathogenesis of severe CHD, targeting them could have a tremendous effect on the most challenging clinical cases.
The Genetic Basis of Maternal Age-Associated Risk of CHD
The maternal age-associated risk of CHD in the Nkx2–5+/− mouse model is a quantitative genetic trait. If the same maternal pathway operates in humans, some offspring may face a higher or lower than chronologically predicted maternal age-associated risk depending on the mother’s genetics. Consideration of biological pathways in the mother that affect the gestational milieu offers potential opportunities to prevent CHD that might not otherwise present themselves by a focus on the embryo.
Insights from each of these areas into the complex genetic basis of CHD could lead to the identification of high-risk populations who would benefit from future prevention strategies. Elucidation of the genetic variants and interactions that affect risk, especially ones that reduce risk, could logically suggest therapies that a focus on monogenic causes has not. One can imagine therapies that target the embryo or the mother depending on where the genes act. The outcomes for newborns who have severe heart defects have improved dramatically in the past few decades, but there is still room for improvement. A prevention strategy, even if only modestly effective, would generate tremendous benefits. In the USA, >30,000 affected children are born each year. The number of adults who have severe CHD is growing by almost 10,000 per year. They exceed the number of children with severe CHD.70 By conservative estimates, the prevention of one case would save around US$100,000 in direct patient care costs and provide society an additional estimated $1,000,000 of economic productivity over a lifetime ($25,000/year×40 years). As little as a 1% reduction or 300 additional, healthy children per year in perpetuity would produce a huge return on investment in research to prevent CHD. Of course, the return on a healthy child for the family is priceless.
Acknowledgments
We thank Drs. Paul Hruz and David Wilson for their comments. E.A. is supported by a NIH predoctoral training grant (T32 HL007873) and a Ruth L. Kirschstein National Research Service Award (F30 HL136077). J.M.N. is supported by an NIH training grant (T32 HD043010, Training of the Pediatric Physician-Scientist). P.Y.J. is an Established Investigator of the American Heart Association and the Lawrence J. & Florence A. DeGeorge Charitable Trust. Additional support was provided by the Children’s Discovery Institute of Washington University and St. Louis Children’s Hospital and the NIH (R01 HL105857).
Footnotes
The authors have no financial disclosures.
References
- 1.Newman JR. The world of mathematics. London: George Allen and Unwin; 1956. [November 1, 2016]. Commentary on Gregor Mendel; pp. 932–936. https://archive.org/details/TheWorldOfMathematicsVolume2. [Google Scholar]
- 2.Peacock TB. On malformations of the human heart. London: John Churchill and Sons; 1866. [November 1, 2016]. pp. 101–166. https://archive.org/details/onmalformationso00peac. [Google Scholar]
- 3.Nora JJ. Multifactorial inheritance hypothesis for the etiology of congenital heart diseases: The genetic-environmental interaction. Circulation. 1968;38:604–617. doi: 10.1161/01.cir.38.3.604. [DOI] [PubMed] [Google Scholar]
- 4.Ferencz C, Neill CA, Boughman JA, Rubin JD, Brenner JI, Perry LW. Congenital cardiovascular malformations associated with chromosome abnormalities: An epidemiologic study. J Pedialr. 1989;114:79–86. doi: 10.1016/s0022-3476(89)80605-5. [DOI] [PubMed] [Google Scholar]
- 5.Hartman RJ, Rasmussen SA, Botto LD, Riehle-Colarusso T, Martin CL, Cragan JD, et al. The contribution of chromosomal abnormalities to congenital heart defects: A population-based study. Pediatr Cardiol. 2011;32:1147–1157. doi: 10.1007/s00246-011-0034-5. [DOI] [PubMed] [Google Scholar]
- 6.Glessner JT, Bick AG, Ito K, Homsy JG, Rodriguez-Murillo L, Fromer M, et al. Increased frequency of de novo copy number variants in congenital heart disease by integrative analysis of single nucleotide polymorphism array and exome sequence data. Circ Res. 2014;115:884–896. doi: 10.1161/CIRCRESAHA.115.304458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zaidi S, Choi M, Wakimoto H, Ma L, Jiang J, Overton JD, et al. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498:220–223. doi: 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Homsy J, Zaidi S, Shen Y, Ware JS, Samocha KE, Karczewski KJ, et al. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science. 2015;350:1262–1266. doi: 10.1126/science.aac9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sifrim A, Hitz MP, Wilsdon A, Breckpot J, Turki SH, Thienpont B, et al. Distinct genetic architectures for syndromic and nonsyndromic congenital heart defects identified by exome sequencing. Nat Genet. 2016;48:1060–1065. doi: 10.1038/ng.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oyen N, Poulsen G, Boyd HA, Wohlfahrt J, Jensen PK, Melbye M. Recurrence of congenital heart defects in families. Circulation. 2009;120:295–301. doi: 10.1161/CIRCULATIONAHA.109.857987. [DOI] [PubMed] [Google Scholar]
- 11.Whittemore R, Wells JA, Castellsague X. A second-generation study of 427 probands with congenital heart defects and their 837 children. J Am Coll Cardiol. 1994;23:1459–1467. doi: 10.1016/0735-1097(94)90392-1. [DOI] [PubMed] [Google Scholar]
- 12.Levy HL, Guldberg P, Guttler F, Hanley WB, Matalon R, Rouse BM, et al. Congenital heart disease in maternal phenylketonuria: Report from the Maternal PKU Collaborative Study. Pediatr Res. 2001;49:636–642. doi: 10.1203/00006450-200105000-00005. [DOI] [PubMed] [Google Scholar]
- 13.Oyen N, Diaz LJ, Leirgul E, Boyd HA, Priest J, Mathiesen ER, et al. Prepregnancy diabetes and offspring risk of congenital heart disease: A nationwide cohort study. Circulation. 2016;133:2243–2253. doi: 10.1161/CIRCULATIONAHA.115.017465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leirgul E, Brodwall K, Greve G, Vollset SE, Holmstrom H, Tell GS, et al. Maternal diabetes, birth weight, and neonatal risk of congenital heart defects in Norway, 1994–2009. Obstet Gynecol. 2016;128:1116–1125. doi: 10.1097/AOG.0000000000001694. [DOI] [PubMed] [Google Scholar]
- 15.Jenkins KJ, Correa A, Feinstein JA, Botto L, Britt AE, Daniels SR, et al. Noninherited risk factors and congenital cardiovascular defects: Current knowledge: A scientific statement from the American Heart Association Council on Cardiovascular Disease in the Young: Endorsed by the American Academy of Pediatrics. Circulation. 2007;115:2995–3014. doi: 10.1161/CIRCULATIONAHA.106.183216. [DOI] [PubMed] [Google Scholar]
- 16.Mefford HC, Sharp AJ, Baker C, Itsara A, Jiang Z, Buysse K, et al. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N Engl J Med. 2008;359:1685–1699. doi: 10.1056/NEJMoa0805384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mazzanti L, Cacciari E. Congenital heart disease in patients with Turner’s syndrome: Italian Study Group for Turner Syndrome (ISGTS) J Pediatr. 1998;133:688–692. doi: 10.1016/s0022-3476(98)70119-2. [DOI] [PubMed] [Google Scholar]
- 18.Chatfield KC, Schrier SA, Li J, Clark D, Kaur M, Kline AD, et al. Congenital heart disease in Cornelia de Lange syndrome: Phenotype and genotype analysis. Am J Med Genet A. 2012;158A:2499–2505. doi: 10.1002/ajmg.a.35582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freeman SB, Bean LH, Allen EG, Tinker SW, Locke AE, Druschel C, et al. Ethnicity, sex, and the incidence of congenital heart defects: A report from the National Down Syndrome Project. Genet Med. 2008;10:173–180. doi: 10.1097/GIM.0b013e3181634867. [DOI] [PubMed] [Google Scholar]
- 20.Newbury-Ecob RA, Leanage R, Raeburn JA, Young ID. Holt-Oram syndrome: A clinical genetic study. J Med Genet. 1996;33:300–307. doi: 10.1136/jmg.33.4.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ryan AK, Goodship JA, Wilson DI, Philip N, Levy A, Seidel H, et al. Spectrum of clinical features associated with interstitial chromosome 22q11 deletions: A European collaborative study. J Med Genet. 1997;34:798–804. doi: 10.1136/jmg.34.10.798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maury P, Gandjbakhch E, Baruteau AE, Bessiere F, Kyndt F, Bouvagnet P, et al. Cardiac phenotype and long-term follow-up of patients with mutations in NKX2–5 Gene. J Am Coll Cardiol. 2016;68:2389–2390. doi: 10.1016/j.jacc.2016.08.064. [DOI] [PubMed] [Google Scholar]
- 23.Rajagopal SK, Ma Q, Obler D, Shen J, Manichaikul A, Tomita-Mitchell A, et al. Spectrum of heart disease associated with murine and human GATA4 mutation. J Mol Cell Cardiol. 2007;43:677–685. doi: 10.1016/j.yjmcc.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Winston JB, Erlich JM, Green CA, Aluko A, Kaiser KA, Takematsu M, et al. Heterogeneity of genetic modifiers ensures normal cardiac development. Circulation. 2010;121:1313–1321. doi: 10.1161/CIRCULATIONAHA.109.887687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bruneau BG, Nemer G, Schmitt JP, Charron F, Robitaille L, Caron S, et al. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell. 2001;106:709–721. doi: 10.1016/s0092-8674(01)00493-7. [DOI] [PubMed] [Google Scholar]
- 26.Meijer WM, Werler MM, Louik C, Hernandez-Diaz S, de Jong-van den Berg LT, Mitchell AA. Can folic acid protect against congenital heart defects in Down syndrome? Birth Defects Res A Clin Mol Teratol. 2006;76:714–717. doi: 10.1002/bdra.20312. [DOI] [PubMed] [Google Scholar]
- 27.Bean LJ, Allen EG, Tinker SW, Hollis ND, Locke AE, Druschel C, et al. Lack of maternal folic acid supplementation is associated with heart defects in Down syndrome: A report from the National Down Syndrome Project. Birth Defects Res A Clin Mol Teratol. 2011;91:885–893. doi: 10.1002/bdra.22848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Digilio MC, Marino B, Capolino R, Angioni A, Sarkozy A, Roberti MC, et al. Familial recurrence of nonsyndromic congenital heart defects in first degree relatives of patients with deletion 22q11.2. Am J Med Genet A. 2005;134A:158–164. doi: 10.1002/ajmg.a.30587. [DOI] [PubMed] [Google Scholar]
- 29.Swaby JA, Silversides CK, Bekeschus SC, Piran S, Oechslin EN, Chow EW, et al. Complex congenital heart disease in unaffected relatives of adults with 22qll .2 deletion syndrome. Am J Cardiol. 2011;107:466–471. doi: 10.1016/j.amjcard.2010.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mlynarski EE, Sheridan MB, Xie M, Guo T, Racedo SE, McDonald-McGinn DM, et al. Copy-number variation of the glucose transporter gene SLC2A3 and congenital heart defects in the 22q11.2 deletion syndrome. Am J Hum Genet. 2015;96:753–764. doi: 10.1016/j.ajhg.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramachandran D, Zeng Z, Locke AE, Mulle JG, Bean LJ, Rosser TC, et al. Genome-wide association study of Down syndrome-associated atrioventricular septal defects. G3 (Bethesda) 2015;5:1961–1971. doi: 10.1534/g3.115.019943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramachandran D, Mulle JG, Locke AE, Bean LJ, Rosser TC, Bose P, et al. Contribution of copy-number variation to Down syndrome-associated atrioventricular septal defects. Genet Med. 2015;17:554–560. doi: 10.1038/gim.2014.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prakash SK, Bondy CA, Maslen CL, Silberbach M, Lin AE, Perrone L, et al. Autosomal and X chromosome structural variants are associated with congenital heart defects in Turner syndrome: The NHLBI GenTAC registry. Am J Med Genet A. 2016;170:3157–3164. doi: 10.1002/ajmg.a.37953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ganguly A, McKnight RA, Raychaudhuri S, Shin BC, Ma Z, Moley K, et al. Glucose transporter isoform-3 mutations cause early pregnancy loss and fetal growth restriction. Am J Physiol Endocrinol Metab. 2007;292:E1241–E1255. doi: 10.1152/ajpendo.00344.2006. [DOI] [PubMed] [Google Scholar]
- 35.Sakata Y, Koibuchi N, Xiang F, Youngblood JM, Kamei CN, Chin MT. The spectrum of cardiovascular anomalies in CHF1/Hey2 deficient mice reveals roles in endocardial cushion, myocardial and vascular maturation. J Mol Cell Cardiol. 2006;40:267–273. doi: 10.1016/j.yjmcc.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 36.Robinson SW, Morris CD, Goldmuntz E, Reller MD, Jones MA, Steiner RD, et al. Missense mutations in CRELD1 are associated with cardiac atrioventricular septal defects. Am J Hum Genet. 2003;72:1047–1052. doi: 10.1086/374319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H, Cherry S, Klinedinst D, Deleon V, Redig J, Reshey B, et al. Genetic modifiers predisposing to congenital heart disease in the sensitized Down syndrome population. Circ Cardiovasc Genet. 2012;5:301–308. doi: 10.1161/CIRCGENETICS.111.960872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, Edie S, Klinedinst D, Jeong JS, Blackshaw S, Maslen CL, et al. Penetrance of congenital heart disease in a mouse model of Down syndrome depends on a trisomic potentiator of a disomic modifier. Genetics. 2016;203:763–770. doi: 10.1534/genetics.116.188045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo T, Chung JH, Wang T, McDonald-McGinn DM, Kates WR, Hawula W, et al. Histone modifier genes alter conotruncal heart phenotypes in 22q11.2 deletion syndrome. Am J Hum Genet. 2015;97:869–877. doi: 10.1016/j.ajhg.2015.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laforest B, Nemer M. GATA5 interacts with GATA4 and GATA6 in outflow tract development. Dev Biol. 2011;358:368–378. doi: 10.1016/j.ydbio.2011.07.037. [DOI] [PubMed] [Google Scholar]
- 41.Maitra M, Schluterman MK, Nichols HA, Richardson JA, Lo CW, Srivastava D, et al. Interaction of Gata4 and Gata6 with Tbx5 is critical for normal cardiac development. Dev Biol. 2009;326:368–377. doi: 10.1016/j.ydbio.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCright B, Lozier J, Gridley T. A mouse model of Alagille syndrome: Notch2 as a gentic modifier of Jag1 haploinsufficiency. Development. 2002;129:1075–1082. doi: 10.1242/dev.129.4.1075. [DOI] [PubMed] [Google Scholar]
- 43.Prall OW, Menon MK, Solloway MJ, Watanabe Y, Zaffran S, Bajolle F, et al. An Nkx2–5/Bmp2/Smad1 negative feedback loop controls heart progenitor specification and prolifieration. Cell. 2007;128:947–959. doi: 10.1016/j.cell.2007.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Santos R, Kawauchi S, Jacobs RE, Lopez-Burks ME, Choi H, Wikenheiser J, et al. Conditional creation and rescue of nipbldeficiency in mice reveals multiple determinants of risk for congenital heart defects. PLoS Biol. 2016;14:e2000197. doi: 10.1371/journal.pbio.2000197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nimura K, Ura K, Shiratori H, Ikawa M, Okabe M, Schwartz RJ, et al. A histone H3 lysine 36 trimethyltransferase links Nkx2–5 to Wolf-Hirschhorn syndrome. Nature. 2009;460:287–291. doi: 10.1038/nature08086. [DOI] [PubMed] [Google Scholar]
- 46.Guris DL, Duester G, Papaioannou VE, Imamoto A. Dose-dependent interaction of Tbx1 and Crkl and locally aberrant RA signaling in a model of del22q11 syndrome. Dev Cell. 2006;10:81–92. doi: 10.1016/j.devcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 47.Racedo SE, McDonald-McGinn DM, Chung JH, Goldmuntz E, Zackai E, Emanuel BS, et al. Mouse and human CRKL is dosage sensitive for cardiac outflow tract formation. Am J Hum Genet. 2015;96:235–244. doi: 10.1016/j.ajhg.2014.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo C, Sun Y, Zhou B, Adam RM, Li X, Pu WT, et al. A Tbx1-Six1/Eya1-Fgf8 genetic pathway controls mammalian cardiovascular and craniofacial morphogenesis. J Clin Invest. 2011;121:1585–1595. doi: 10.1172/JCI44630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vitelli F, Zhang Z, Huynh T, Sobotka A, Mupo A, Baldini A. Fgf8 expression in the Tbx1 domain causes skeletal abnormalities and modifies the aortic arch but not the outflow tract phenotype of Tbx1 mutants. Dev Biol. 2006;295:559–570. doi: 10.1016/j.ydbio.2006.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vincent SD, Mayeuf-Louchart A, Watanabe Y, Brzezinski JA, Miyagawa-Tomita S, Kelly RG, et al. Prdm1 functions in the mesoderm of the second heart field, where it interacts genetically with Tbxl, during outflow tract morphogenesis in the mouse embryo. Hum Mol Genet. 2014;23:5087–5101. doi: 10.1093/hmg/ddu232. [DOI] [PubMed] [Google Scholar]
- 51.Winston JB, Schulkey CE, Chen IB, Regmi SD, Efimova M, Erlich JM, et al. Complex trait analysis of ventricular septal defects caused by Nkx2 – 5 mutation. Circ Cardiovasc Genet. 2012;5:293–300. doi: 10.1161/CIRCGENETICS.111.961136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De LA, Sarkozy A, Consoli F, Ferese R, Guida V, Dentici ML, et al. Familial transposition of the great arteries caused by multiple mutations in laterality genes. Heart. 2010;96:673–677. doi: 10.1136/hrt.2009.181685. [DOI] [PubMed] [Google Scholar]
- 53.Granados-Riveron JT, Pope M, Bu’lock FA, Thornborough C, Eason J, Setchfield K, et al. Combined mutation screening of NKX2–5, GATA4, and TBX5 in congenital heart disease: Multiple heterozygosity and novel mutations. Congenit Heart Dis. 2012;7:151–159. doi: 10.1111/j.1747-0803.2011.00573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Topf A, Griffin HR, Glen E, Soemedi R, Brown DL, Hall D, et al. Functionally significant, rare transcription factor variants in tetralogy of Fallot. PLoS One. 2014;9:e95453. doi: 10.1371/journal.pone.0095453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.D’Alessandro LC, Al Turki S, Manickaraj AK, Manase D, Mulder BJ, Bergin L, et al. Exome sequencing identifies rare variants in multiple genes in atrioventricular septal defect. Genet Med. 2016;18:189–198. doi: 10.1038/gim.2015.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li Y, Yagi H, Onuoha EO, Damerla RR, Francis R, Furutani Y, et al. DNAH6 and its interactions with PCD genes in heterotaxy and primary ciliary dyskinesia. PLoS Genet. 2016;12:e1005821. doi: 10.1371/journal.pgen.1005821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Y, Klena NT, Gabriel GC, Liu X, Kim AJ, Lemke K, et al. Global genetic analysis in mice unveils central role for cilia in congenital heart disease. Nature. 2015;521:520–524. doi: 10.1038/nature14269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jay PY, Akhirome E, Magnan RA, Zhang MR, Kang L, Qin Y, et al. Transgenerational cardiology: One way to a baby’s heart is through the mother. Mol Cell Endocrinol. 2016;435:94–102. doi: 10.1016/j.mce.2016.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Forrester MB, Merz RD. Descriptive epidemiology of selected congenital heart defects, Hawaii, 1986 – 1999. Paediatr Perinat Epidemiol. 2004;18:415–424. doi: 10.1111/j.1365-3016.2004.00594.x. [DOI] [PubMed] [Google Scholar]
- 60.Hollier LM, Leveno KJ, Kelly MA, MCIntire DD, Cunningham FG. Maternal age and malformations in singleton births. Obstet Gynecol. 2000;96:701–706. doi: 10.1016/s0029-7844(00)01019-x. [DOI] [PubMed] [Google Scholar]
- 61.Kidd SA, Lancaster PA, McCredie RM. The incidence of congenital heart defects in the first year of life. J Paediatr Child Health. 1993;29:344–349. doi: 10.1111/j.1440-1754.1993.tb00531.x. [DOI] [PubMed] [Google Scholar]
- 62.Materna-Kiryluk A, Wisniewska K, Badura-Stronka M, Mejnartowicz J, Wieckowska B, Balcar-Boron A, et al. Parental age as a risk factor for isolated congenital malformations in a Polish population. Paediatr Perinat Epidemiol. 2009;23:29–40. doi: 10.1111/j.1365-3016.2008.00979.x. [DOI] [PubMed] [Google Scholar]
- 63.Miller A, Riehle-Colarusso T, Siffel C, Frias JL, Correa A. Maternal age and prevalence of isolated congenital heart defects in an urban area of the United States. Am J Med Genet A. 2011;155A:2137–2145. doi: 10.1002/ajmg.a.34130. [DOI] [PubMed] [Google Scholar]
- 64.Pradat P, Francannet C, Harris JA, Robert E. The epidemiology of cardiovascular defects, part I: A study based on data from three large registries of congenital malformations. Pediatr Cardiol. 2003;24:195–221. doi: 10.1007/s00246-002-9401-6. [DOI] [PubMed] [Google Scholar]
- 65.Reefhuis J, Honein MA. Maternal age and non-chromosomal birth defects, Atlanta 1968 – 2000: Teenager or thirty-something, who is at risk? Birth Defects Res A Clin Mol Teratol. 2004;70:572–579. doi: 10.1002/bdra.20065. [DOI] [PubMed] [Google Scholar]
- 66.Schulkey CE, Regmi SD, Magnan RA, Danzo MT, Luther H, Hutchinson AK, et al. The maternal-age-associated risk of congenital heart disease is modifiable. Nature. 2015;520:230–233. doi: 10.1038/nature14361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kuyumcu-Martinez MN, Bressan MC. Rebuilding a broken heart: Lessons from developmental and regenerative biology. Development. 2016;143:3866–3870. doi: 10.1242/dev.143842. [DOI] [PubMed] [Google Scholar]
- 68.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hoffman JI, Kaplan S. The incidence of congenital heart disease. J Am Coll Cardiol. 2002;39:1890–1900. doi: 10.1016/s0735-1097(02)01886-7. [DOI] [PubMed] [Google Scholar]
- 70.Gilboa SM, Devine OJ, Kucik JE, Oster ME, Riehle-Colarusso T, Nembhard WN, et al. Congenital heart defects in the United States: Estimating the magnitude of the affected population in 2010. Circulation. 2016;134:101–109. doi: 10.1161/CIRCULATIONAHA.115.019307. [DOI] [PMC free article] [PubMed] [Google Scholar]