Fig. 3.
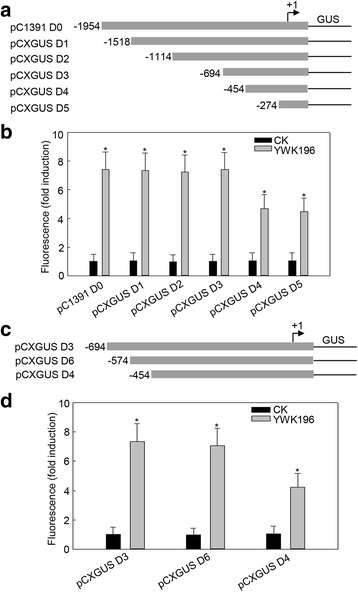
Quantitative fluorometric assays for GUS activity driven by various GRMZM2G174449 promoter deletion constructs. a Diagram of various deletion derivatives of the GRMZM2G174449 promoter. All promoter derivatives were fused to a GUS reporter gene. b The GUS activity of the DNA constructs in a NB transient expression system. The NB leaves were inoculated with R. solani strain YWK196 for 24 h. GUS activity was calculated relative to CK of the pC1391 D0 construct. Asterisks indicate statistically significant differences, as determined by Student’s t-tests (*P < 0.05). c Diagram of the deletion constructs in the −694 to −455 region of the GRMZM2G174449 promoter. All promoter derivatives were fused to a GUS reporter gene. d The GUS activity of DNA constructs prepared in a NB leaf transient expression system. The NB leaves were inoculated with R. solani strain YWK196 for 24 h. GUS activity was calculated relative to CK of the pCXGUS D3 construct. Asterisks indicate statistically significant differences, as determined by Student’s t-tests (*P < 0.05)
