Fig. 1.
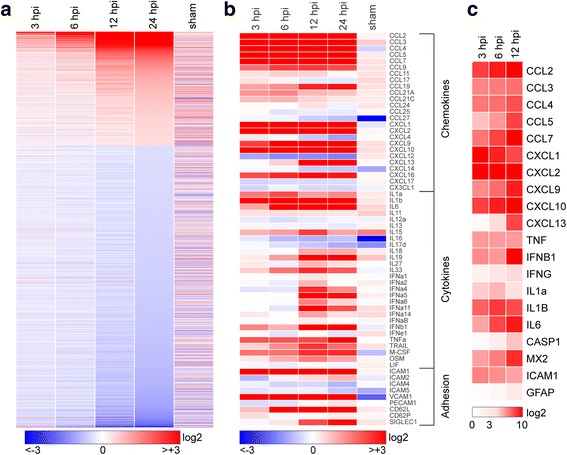
Transcriptional program initiated in the hippocampus by acute infection. Hippocampal tissue was collected for RNA purification at early timepoints following intracranial inoculation of adult female C57Bl/6 mice with 2 × 105 PFU of the Daniel’s strain of TMEV. Transcriptional changes were analyzed using Illumina MouseWG-6 v2.0 beadchips. Fold change relative to uninfected control for 16,900 genes present on the array was calculated, converted to log2, and heatmapped using Gitools. Sham controls were injected intracranially with culture media used for the virus preparation, and RNA was collected at 24 h postinfection (hpi). Expression data for all conditions except sham are the means from six individual mice analyzed in two separate experiments (three mice per experiment). Three sham mice were analyzed in the second experiment. a Heatmap showing pattern of transcriptional changes at 3, 6, 12, and 24 hpi relative to uninfected controls. The discontinuity in expression coding is the result of removing from analysis all genes with changes between − 1.2-fold and + 1.2-fold at 24 hpi, leaving 6764 genes in the heatmap. Color-coded heatmaps range from greater than eightfold downregulated (log2 < − 3.0; blue) to greater than eightfold upregulated (log2 > + 3.0; red). b The same expression data remapped to highlight chemokines, cytokines, and adhesion factors. In contrast to (a), values are continuous between negative eightfold and positive eightfold. Note that the color range was selected to reveal weaker changes in some genes, resulting in saturation at all timepoints for highly upregulated genes such as CCL2. c A further subset of highly upregulated and notable genes was validated by RT-PCR and the results heatmapped on a scale from 0 to 1000-fold (log2 = + 10) to reveal large expression changes over the first 12 h
